Details of the Drug
General Information of Drug (ID: DMRXPFM)
| Drug Name |
BZ5
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Indication |
|
||||||||||||||||||||||
| Structure |
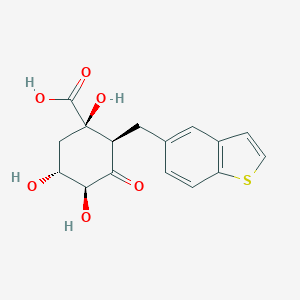 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 336.4 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 0.6 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 3 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 4 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 7 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Interaction Atlas (MIA) | |||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| The Studied Disease | Discovery agent | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICD Disease Classification | N.A. | |||||||||||||||||||||||
|
||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||
