Keywords Search
Entries List Search
Drug 2D Structure Similarity Search
Molecular Sequence Similarity Search
Drug Information in DrugMAP
Drug Off-Target (DOT) Information in DrugMAP New
Disease / Drug Indication Information in DrugMAP New
Drug Therapeutic Target (DTT) Information in DrugMAP
Drug Transporter (DTP) Information in DrugMAP
Drug-Metabolizing Enzyme (DME) Information in DrugMAP
Search Methods Available in DrugMAP
Data Content Available in DrugMAP
General Information of Drug
Molecular Interaction Atlas of Drug
Molecular Expression Atlas of Drug
Drug-Drug Interaction (DDI) Information of Drug
Pharmaceutical Excipient & Formulation Information of Drug
Drug Combination (DrugComb) Profiles of Drug New
Drug Combination Information in DrugMAP New
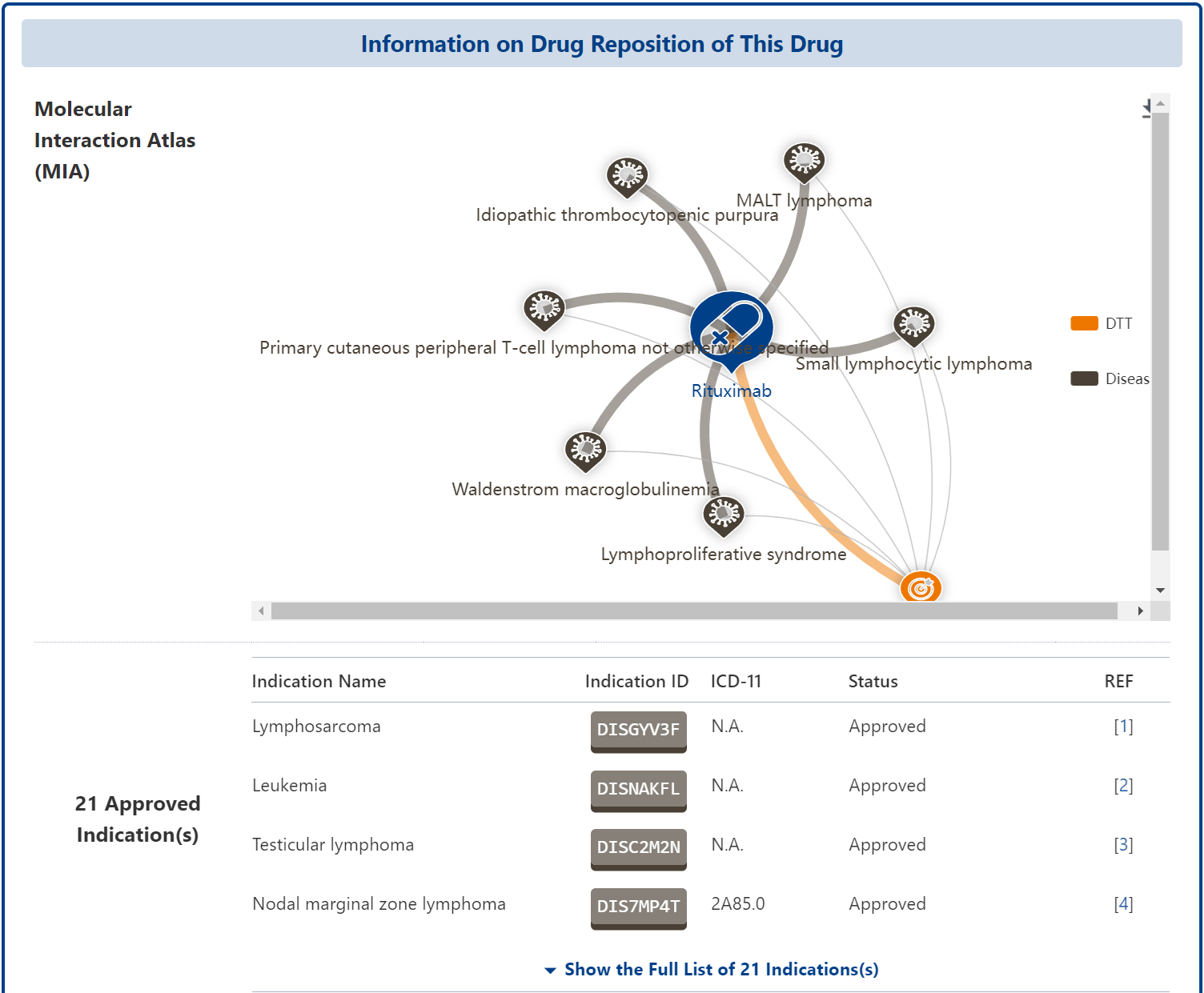
Drug Reposition (DrugRepo) Profiles of Drug New
General Information of DOT New
Related Disease Information of DOT New
Molecular Interaction Atlas of DOT New
General Information of Disease New
Drug-Interaction Atlas (DIA) of Disease New
Molecular Interaction Atlas (MIA) of Disease New
General Information of DTT
Molecular Interaction Atlas of DTT
Molecular Expression Atlas of DTT
Information on Pharmacokinetic Roles of DTT
General Information of DTP
Molecular Interaction Atlas of DTP
Molecular Expression Atlas of DTP
Drug Affinity Data of DTP
Information on Pharmacodynamic Roles of DTP
General Information of DME
Molecular Interaction Atlas of DME
Molecular Expression Atlas of DME
Experimental DME Kinetic Data of Drug
Information on Pharmacodynamic Roles of DME
The most frequently used keyword searches are set up at the top of the "Home" page of DrugMAP and each search page. In the keyword search, users can search for the entries they wanted using any keyword related to the entries they were searching for.

Taking a drug as an example, the users can search and find their wanted drugs by entering the keywords about drug name, drug synonyms, drug PubChem ID, drug target, drug-metabolizing enzyme, drug transporter and any other keywords related to the drug.
Query can be submitted by entering keywords into the main searching frame. Users can specify part or full name in the text field. To facilitate a more customized input query, the wildcard characters of "*", "-" and "?" are also supported in DrugMAP. For example:
If search "Estradiol Acetate" and click the "Drug" button, find three entries with name "Estradiol", "Estradiol Acetate" and "Segesterone acetate; ethinyl estradiol". If you only want the result of "Estradiol Acetate", double quotation marks are required when you search the drug name;
If search: "Alpha-1? adrenergic receptor", finds entries with target names like "Alpha-1A adrenergic receptor", "Alpha-1B adrenergic receptor" or "Alpha-1C adrenergic receptor" etc. Here, "?" represents any one character;
If search: "Alpha-* adrenergic receptor", finds the same entries as above. Here "*" represents a string of any length. In this case, it represents "1A", "1B" or "1C" respectively.
In addition to the usual keyword searches, DrugMAP also offers Entries List-based searches for Entries such as Drug, Target, Drug Transporter (DT), drug-metabolizing enzyme (DME), etc. The main difference between this List-based search and the keyword search is that the List-based search matches the content text in an exact rather than a fuzzy way, in other words, only content that is identical to the entry selected by the user will be retrieved.

In each of the different drop-down search boxes, DrugMAP provides information about different Entries, for example, in the case of Drug, the 5 drop-down menus related to drug provide information about the drug's "Drug Name", "ICD-11 Defined Disease Class", "Target Name", "DT Name", and "DME Name" , the user can select the item they are interested in to search for drugs related to this item.
In addition to the two text-based search methods mentioned above, a 2D structural similarity-based search specifically for drugs was also provided in DrugMAP. Users can click the "Tanimoto-based Chemical Structure Similarity Search" block to use this search method

On this search page, users can retrieve drugs with similar 2D structures to those entered by (1) manually drawing the 2D structure of the drug on the JSME molecular editor; (2) uploading a local 2D structure file of the drug in SDF/MOL format; and (3) entering the SMILES string of the chemical structure.
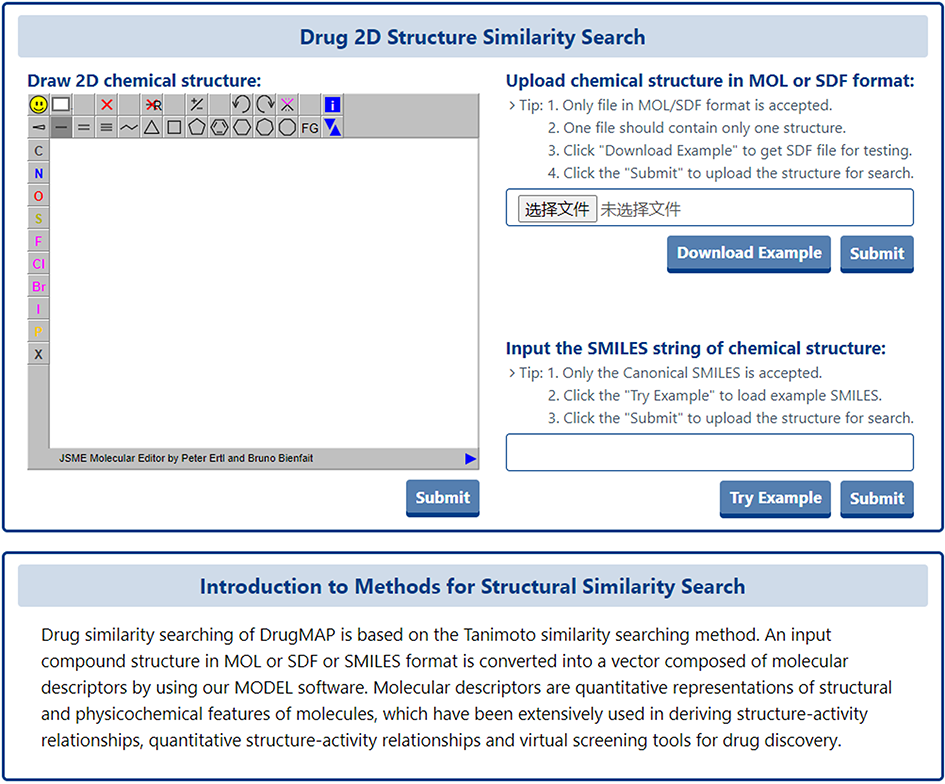
Drug similarity searching of DrugMAP is based on the Tanimoto similarity searching method. An input compound structure in MOL or SDF or SMILES format is converted into a vector composed of molecular descriptors by using our MODEL software. Molecular descriptors are quantitative representations of structural and physicochemical features of molecules, which have been extensively used in deriving structure-activity relationships, quantitative structure-activity relationships and virtual screening tools for drug discovery.
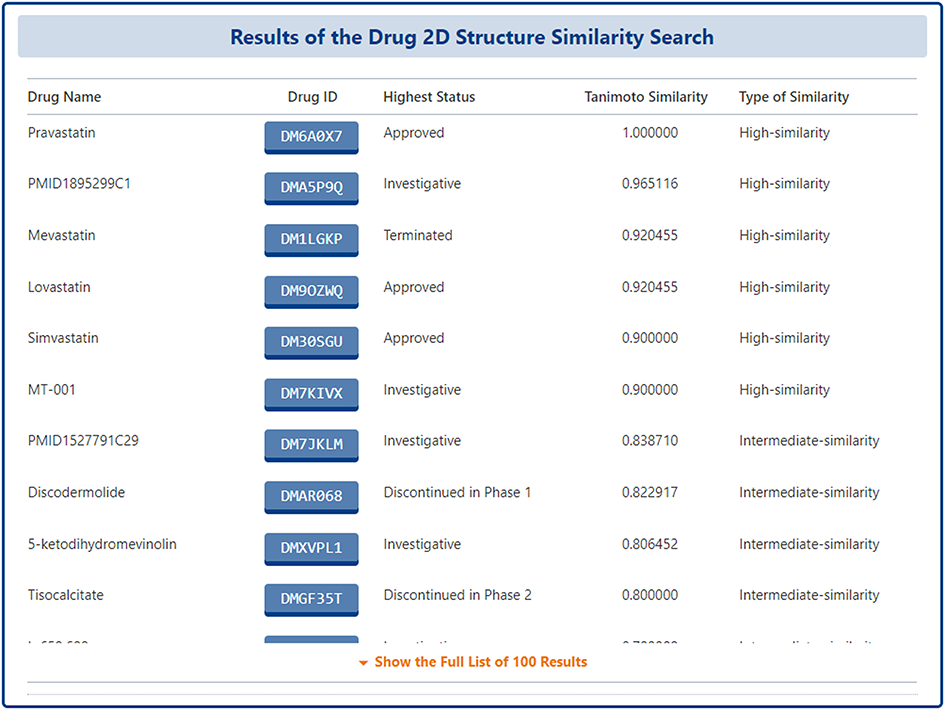
On the search results page, DrugMAP lists the top-100 drugs that are most similar to the 2D structure entered by the user in a table, and the drugs are ranked according to their Tanimoto Similarity scores, with the drug's "Drug Name", "Drug ID", "Highest Status", "Tanimoto Similarity" and the "Type of Similarity" information is listed directly on the search results page, and users can click on the Drug ID to go to the details page.
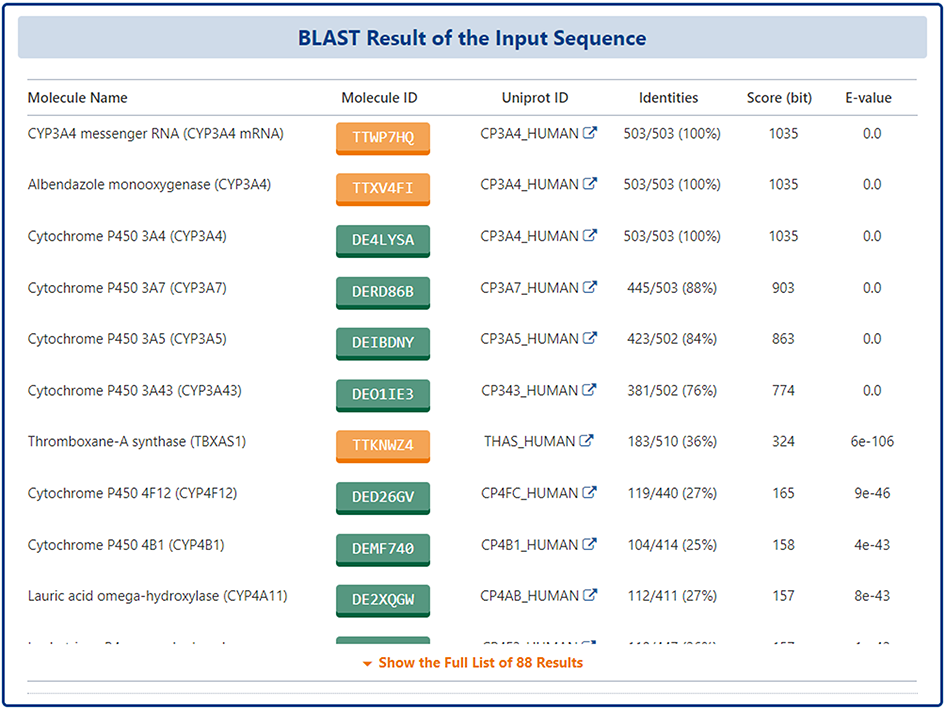
For the molecule of pharmaceutical importance (DTT, DTP, DME), a BLASTp-based molecular sequence similarity search method is also provided in DrugMAP. Users can click the "BLAST-based Molecular Sequence Similarity Search" block on the home page to use this search method.

On this page, the user can enter 2D sequence in FASTA format of any protein and submit it, and DrugMAP will perform a sequence-based protein similarity search across all types of molecules (DTT, DTP, DME).
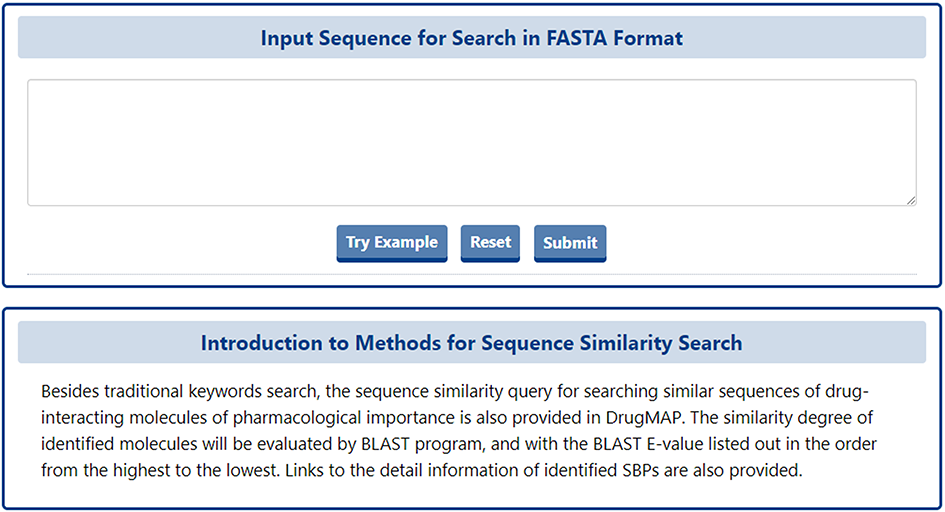
The similarity degree of identified molecules will be evaluated by BLAST program, and with the BLAST E-value listed out in the order from the highest to the lowest. Links to the detail information of identified SBPs are also provided.
On the search results page, DrugMAP lists molecules with significant similarity to the sequence entered by the user, all molecules are sorted by E-value size, and the results table provides "Molecule Name", " Molecule ID", "UniPort ID", "Identities", "Score (bit)", "E-value", etc. The user can click on the Molecule ID to access the molecule details page.
On the search page for drugs, DrugMAP offers a keyword search field for drugs and 5 drop down menu search fields, the 5 drop-down menus related to drug provide information about the drug's "Drug Name", "ICD-11 Defined Disease Class", "Target Name", "DT Name", and "DME Name", the user can select the item they are interested in to search for drugs related to this item.

The search results page for a drug briefly displays information about Drug Name, Drug Status, Indication, and Related Molecule(s), specifically, in the "elated Molecule(s)" section, three different logo colours show the presence of Target, DT and DME information is present or not. And the "Molecular Interaction Atlas" and the "Molecular Expression Atlas" buttons directly linked to the molecular interaction/expression atlas page of this drug.
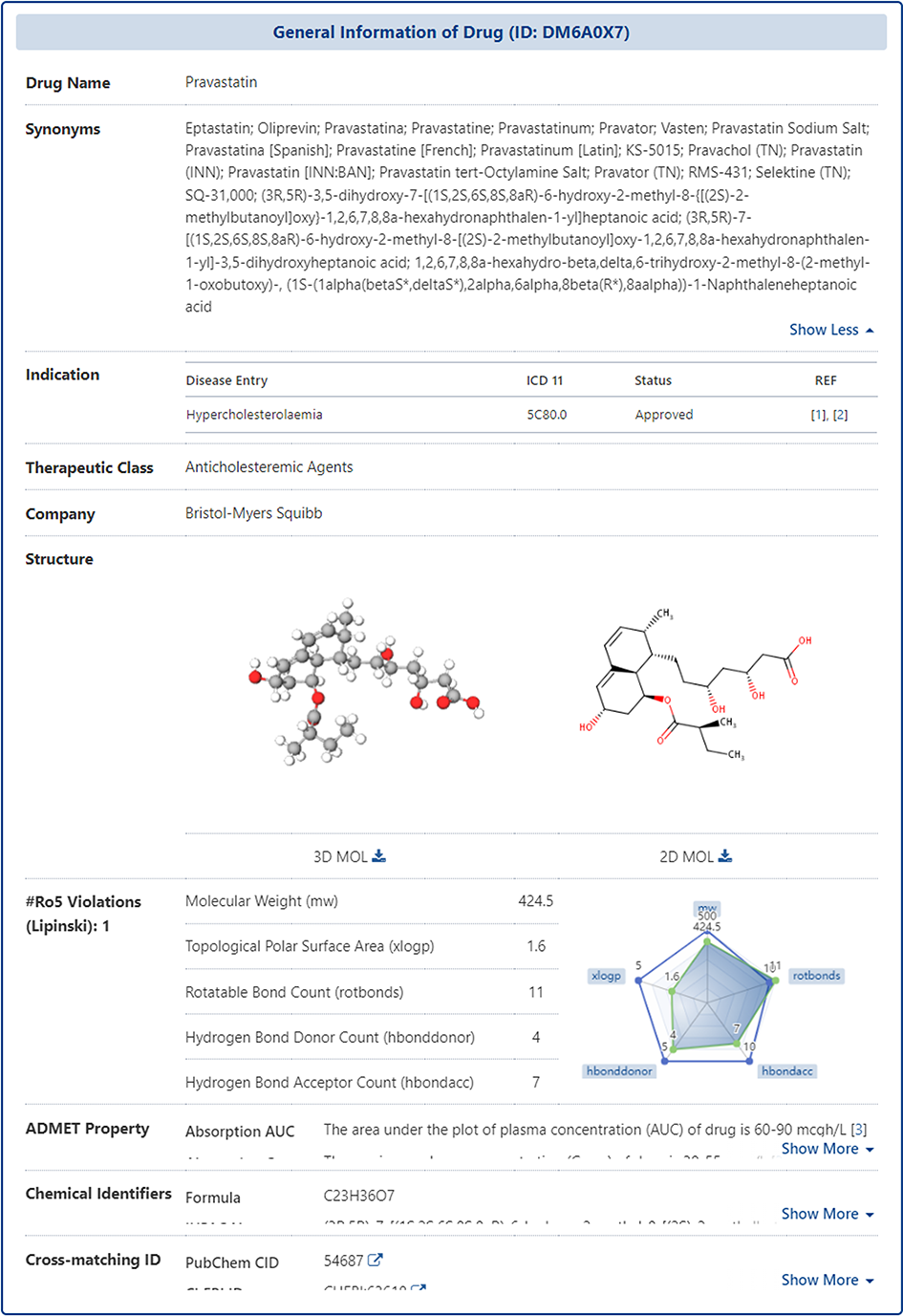
In the "General Information of Drug" section of the Drug Information page, DrugMAP provides information on the Name, Synonyms, Indication, Therapeutic Class, Structure, Lipinski Ro5 Violations, ADMET Property (Absorption, Distribution, Metabolism, Elimination, etc.), Chemical Identifiers (Formula, IUPAC Name, Canonical SMILES, etc.), Cross-matching ID (PubChem CID, ChEBI ID, CAS Number, etc.).
Molecular Interaction Atlas of Drug
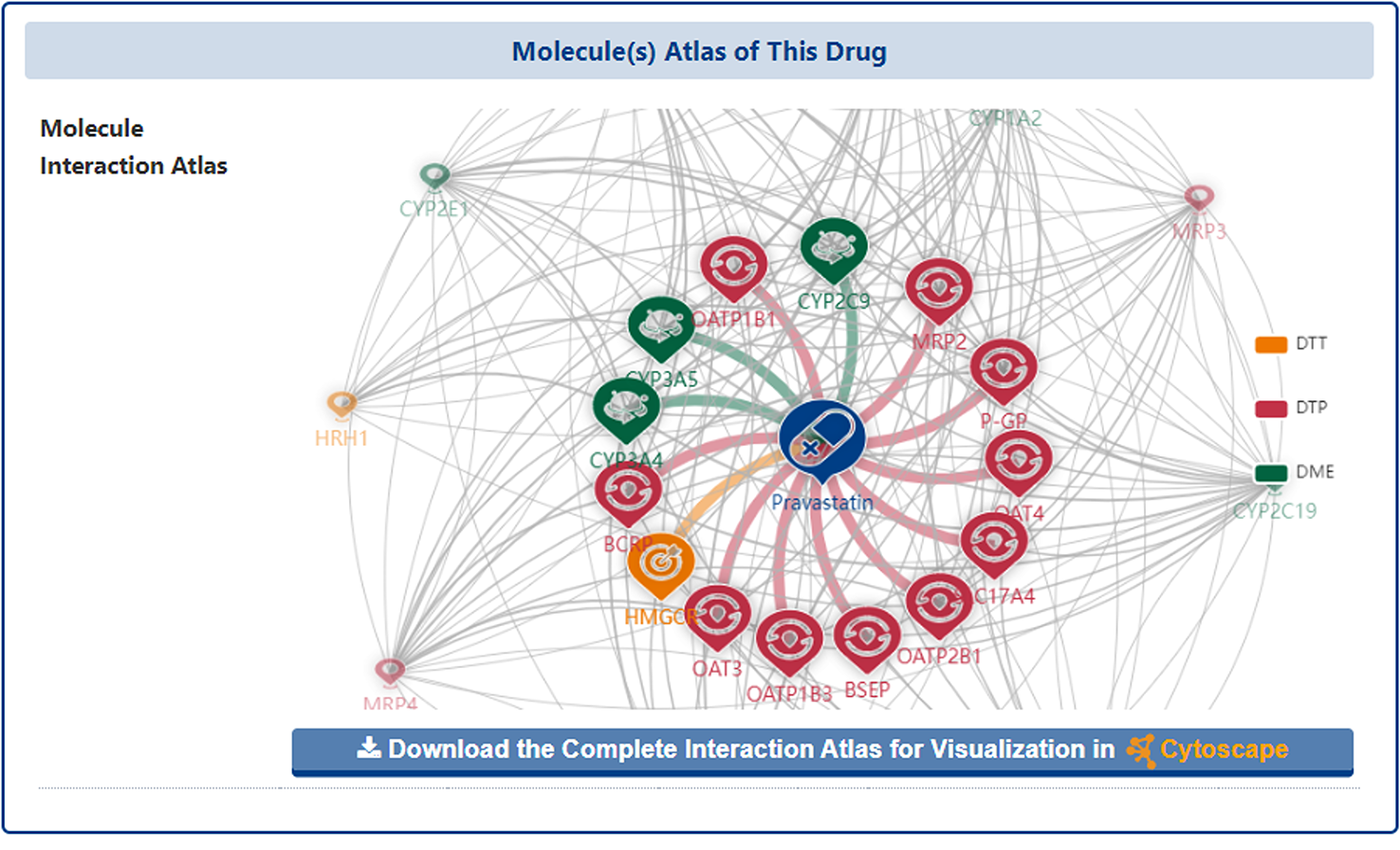
In the "Molecular Interaction Atlas of This Drug" section of the drug information page, DrugMAP provides basic information about all the molecule(s) of pharmaceutical importance that interacting with the drug (including DTT, DTP and DME) in form of interaction tables and map.
In the interaction table, the name, ID and UniProt ID of all molecules and the Highest Status of their interacting drugs are listed, and users can click on the name of DTT, DTP and DME to access the information page of DTT, DTP and DME.
In the interaction map, the drug is placed right in the centre and all molecules that interact directly with the drug (directly-interacted molecules) are distributed around and directly connected to the drug. In addition, the other molecules that have shared drugs with the directly-interacted molecules are represented by small translucent landmarks and are scattered around the map, with a grey line between two molecules representing that there were shared interacting drug between them.
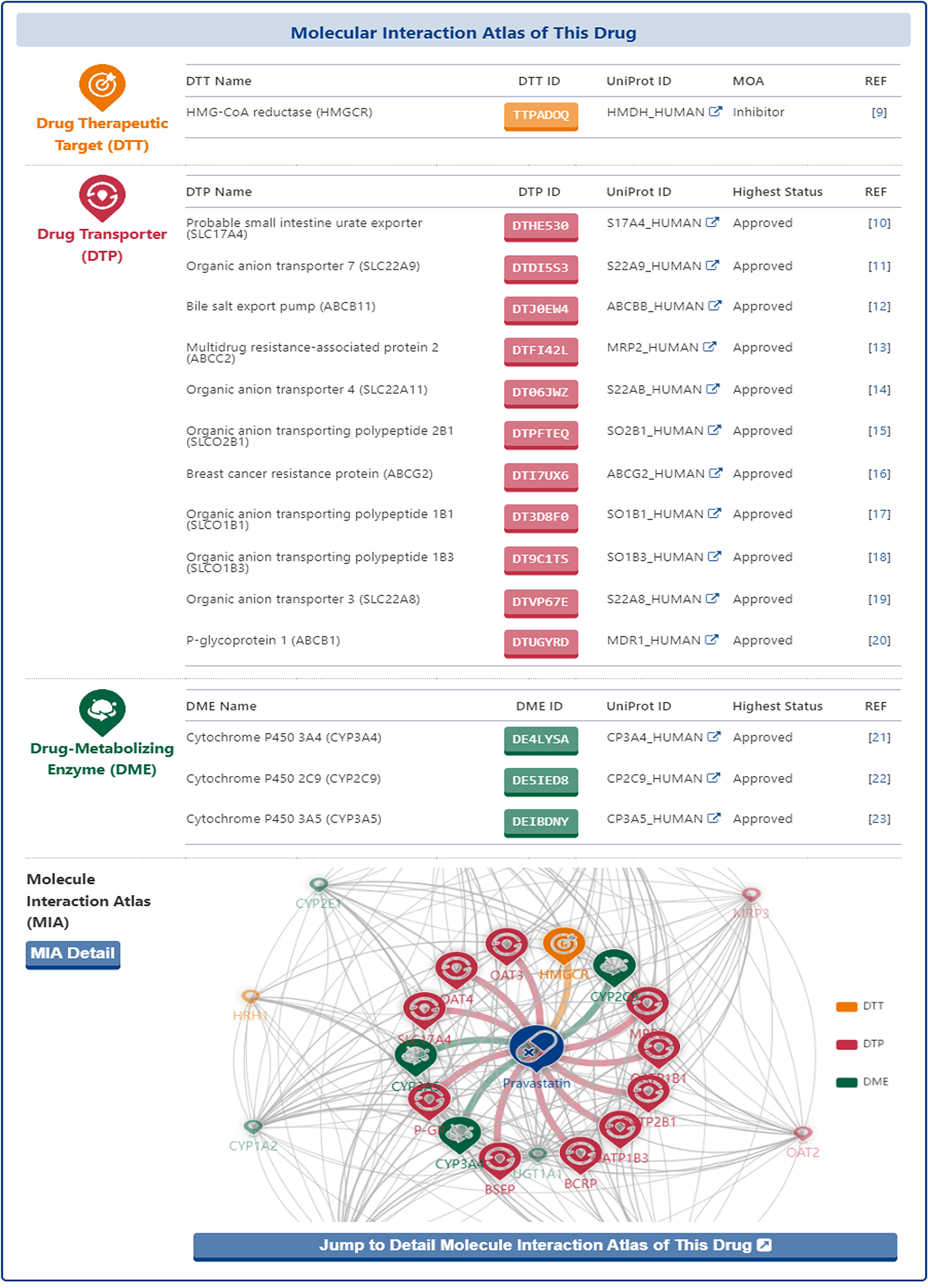
Moreover, the users could click the "Jump to the Detail Molecular Interaction Atlas of This Drug" and "MIA Detail" buttons to jump to the "Detail Molecular Interaction Atlas of Drug" page.
In the "Detail Molecular Interaction Atlas of Drug" page, all molecules that interact directly with the subject drug and the other drugs they interact with are presented in the form of a sankey diagram, in which the first layer is the subject drug, the second layer is the classification of molecules (i.e. DTT, DTP, DME), the third layer is the molecules that interact directly with the subject drug and finally, the fourth layer is all other drugs that interact with molecules. The interactions between the layers are shown by the gradient-coloured curved lines. The Sankey diagram is interactive and the user can mouse over the node at each level to see the details of that node.
In addition to this, all the drugs interacting with each different molecule are listed in a table at the bottom of the Sankey diagram, where the user can see the "Drug Name", "Drug ID", "Indication", "ICD-11", "Highest Status", or click on the ID of each drug to access its details page.
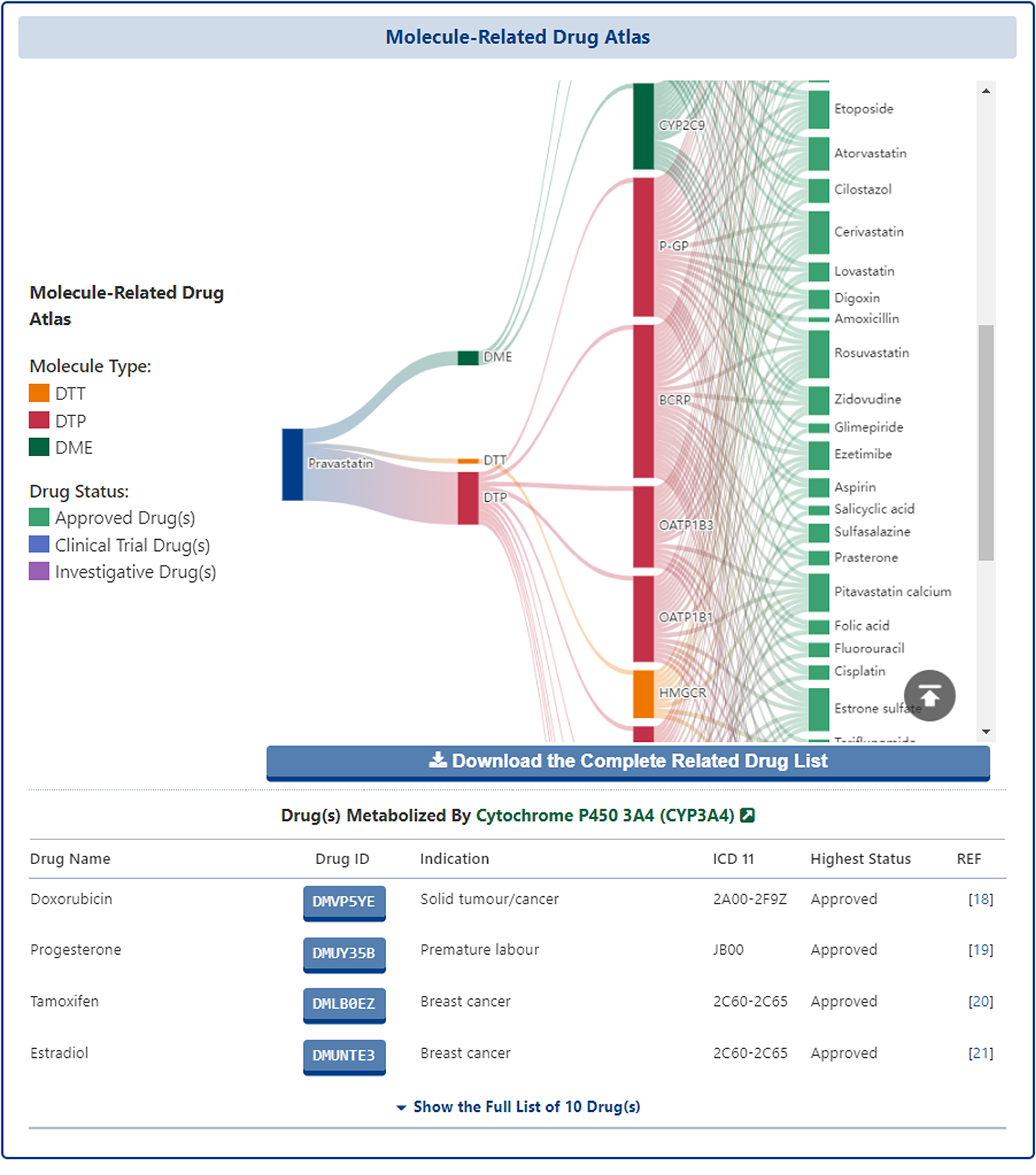
At the bottom of the page, DrugMAP re-presents the Molecule Interaction Atlas and provides a "Download the Complete Interaction Atlas for Visualization in Cytoscape" download button, which allows users to download all the interaction network data of the subject drug for further network analysis and visualization locally or on Cytoscape.
Molecular Expression Atlas of Drug
In addition to the above sections, DrugMAP also provides the expression atlas of the molecules associated with this drug.
In the section of "Molecular Expression Atlas of This Drug", DrugMAP collected expression microarray data from healthy individual and patients associated with the indication of the drug in focal tissue and calculated fold change, p-value and Z-score, and finally presented the data in box plots and tables.
In the expression table, DrugMAP provides p-value, Fold-Change and Z-score values for the differential expression of molecules that interact directly with the subject drug between focal and healthy tissues, while in the graph below the table, the differential expression of molecules between focal and healthy tissues is presented in a more visual and interactive way. The solid green dots represent healthy tissue samples, while the solid red dots represent diseased tissue samples, and the user can mouse over the solid dot to see the GSM ID and normalized expression values for each sample, or mouse over the box to see the p-value, Fold-Change and Z-score.
The data used in the Molecular Expression Atlas is derived from a collection of 5,534 series records of human gene expression raw data. These records are based on the Affymetrix HGU133 Plus 2.0 platform and were obtained from the GEO database. Disease and tissue distributions were identified from the sample annotations associated with these records. To illustrate the expression atlas of all drug-interacting molecules, a careful selection process was undertaken. The disease indications of drugs were compared to the collected records, resulting in the selection of 436 records. In cases where there were small sample sizes, multiple records from the same disease and tissue were integrated to address low statistical power. The collected records underwent rigorous pre-processing to ensure reliable and standardized data analysis. This involved normalization, log transformation, data integration, perfect match correction, quantization, powerful multi-array averaging, and median polishing.
Differential expression between distinct groups of samples (disease tissues of patients vs. normal tissues of healthy individuals) was assessed using various statistical methods. Fold change, P value, and Z score were calculated to determine the significance of differential expression. However, it should be noted that DrugMAP used the integrated GEO data for analysis. The p-value provides a direct measure of the significance of gene expression differences, while the adj p-value, which takes into account multiple test correction, may sometimes be too conservative, resulting in false negative results. Therefore, the adjusted p-value is not used in this analysis.The calculation methods for fold change, P value and Z score are as follows:
Fold change: calculated by comparing the ratio of gene expression levels in different sample groups (e.g. diseased tissue vs. healthy tissue).
p-value: calculated using Student's t-test to assess the statistical significance of the difference between the two sample groups.
Z score: is calculated from the standardized data and indicates the deviation of a value from the mean, expressed in standard deviations. In RNA-seq and microarray studies, tools such as limma are often used to perform these statistical calculations.
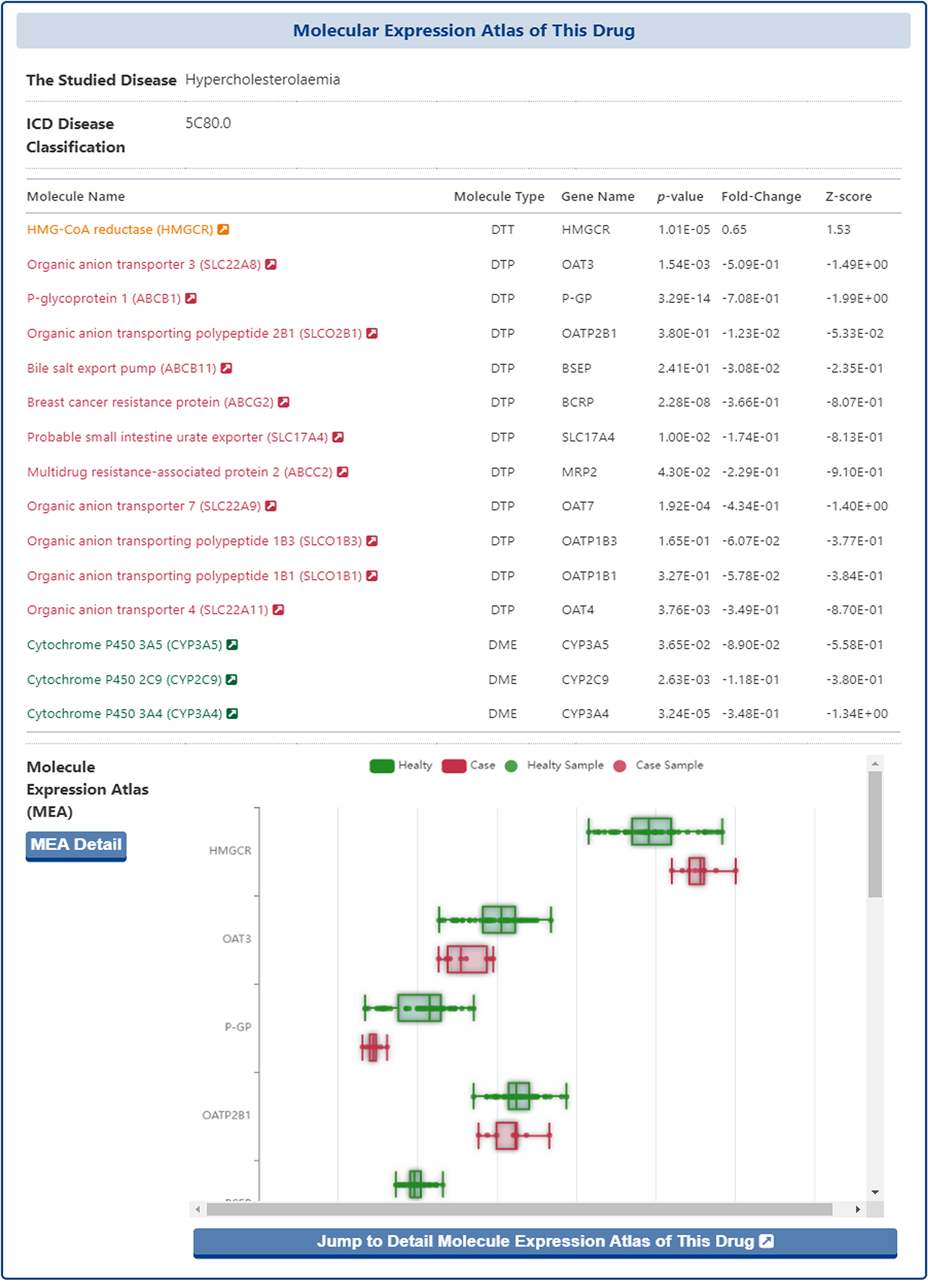
Moreover, the users could click the "Jump to the Detail Molecular Expression Atlas of This Drug" and "MIA Detail" buttons to jump to the "Detail Molecular Expression Atlas of Drug" page.
In the "Detail Molecular Expression Atlas of Drug" page, DrugMAP provides a comparison of the expression of molecules that directly interact with the subject drug in four ADME-related organs (Liver, Colon, Kidney, Small Intestine) in the form of a table and a bar chart.
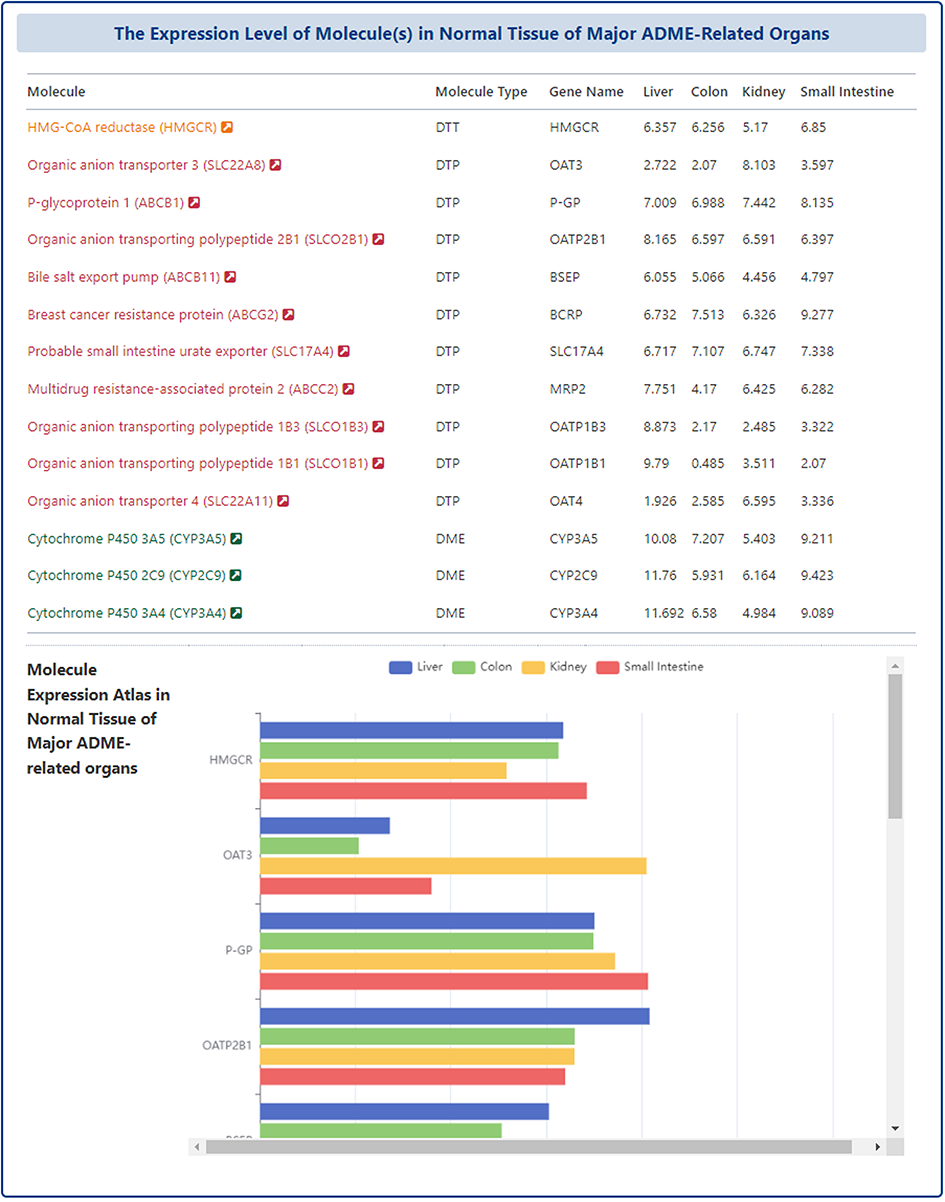
Drug-Drug Interaction (DDI) Information of Drug
In addition to drug-molecule interactions, DrugMAP also collected information on drug-drug interactions (DDI) related to the subject drug.
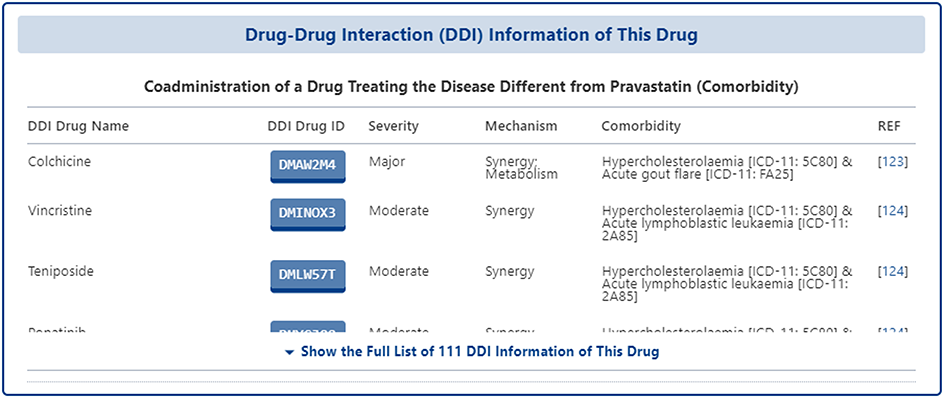
In the section of "Drug-Drug Interaction (DDI) Information of This Drug", all of the DDI information related to the subject drug are collected from FDA label, Drugs.com, literature research, and other reliable information sources. The data collected are presented in a table format and for each DDI, DrugMAP provides information such as "DDI Drug Name", "DDI Drug ID", "Severity", "Mechanism", "Comorbidity", "Reference", etc. "Reference" and other information.
Pharmaceutical Excipient & Formulation Information of Drug
The way in which a drug is formulated can play a pivotal role in the ADMET of a drug and can therefore also have a huge impact on the final efficacy of the drug. Information on drug formulations and drug excipients is essential for a complete understanding of the ADMET of a drug. For this reason, DrugMAP provides comprehensive information on drug formulations and drug excipients.
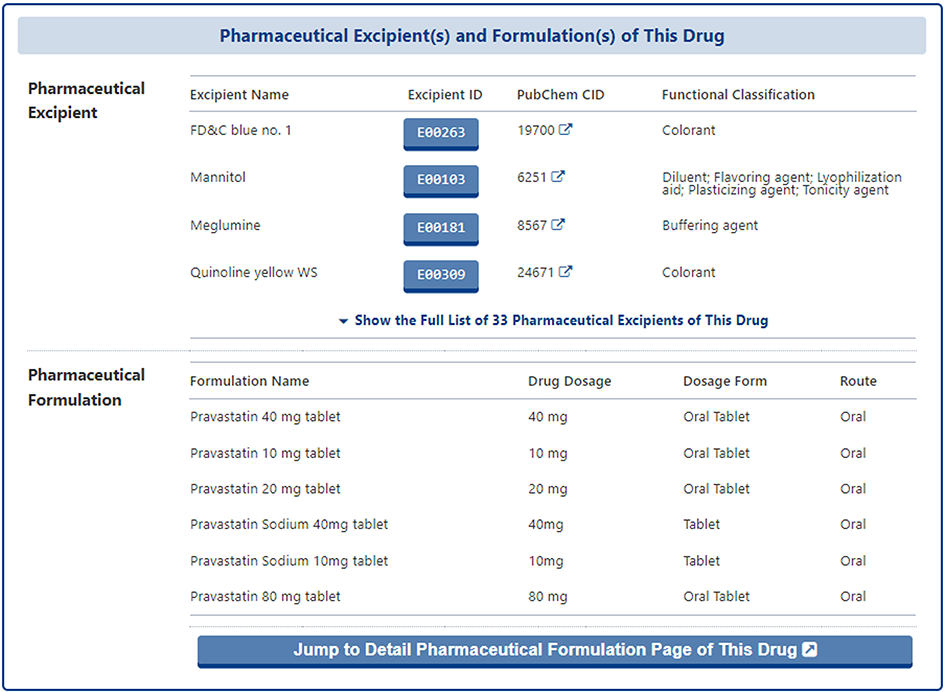
In the section of "Pharmaceutical Excipient(s) and Formulation(s) of This Drug", the pharmaceutical excipient that co-administered the subject drug and the pharmaceutical formulation that contained the subject drug are provided in two separate tables. In the table of pharmaceutical excipient, the "Excipient Name", "Excipient ID", "PubChem ID", "Functional Classification" of excipient are provided. And in the table of pharmaceutical formulation, the formulation information including "Formulation Name", "Drug Dosage", "Dosage Form", and "Route" of the subject drug are provided.
In addition, users can click the "Jump to the Detail Pharmaceutical Formulation Page of This Drug" to access the detail page of pharmaceutical formulation.
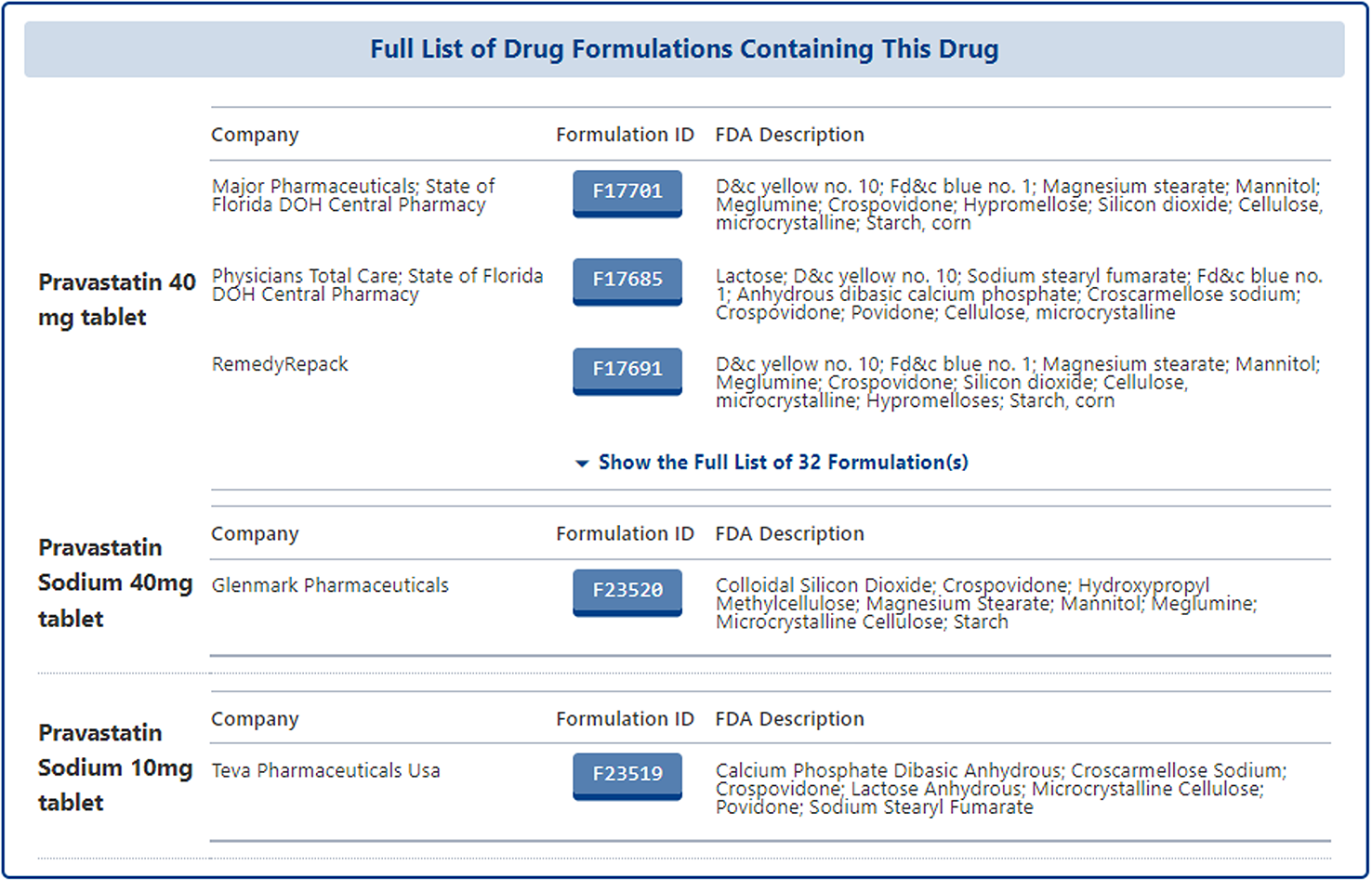
In the detail page of pharmaceutical formulation of subject drug, all formulations related to the subject drug from different companies are listed with the following information: "Company", "Formulation ID", "FDA Description". Users can click the "Formulation ID" to access the detail page of each specific formulation.
In the detail page of each specific formulation, users can find information on the active pharmaceutical ingredient" and the full list of all pharmaceutical excipients. If the users are interested in one of the excipient, they can also click on the Excipient ID to access its details page
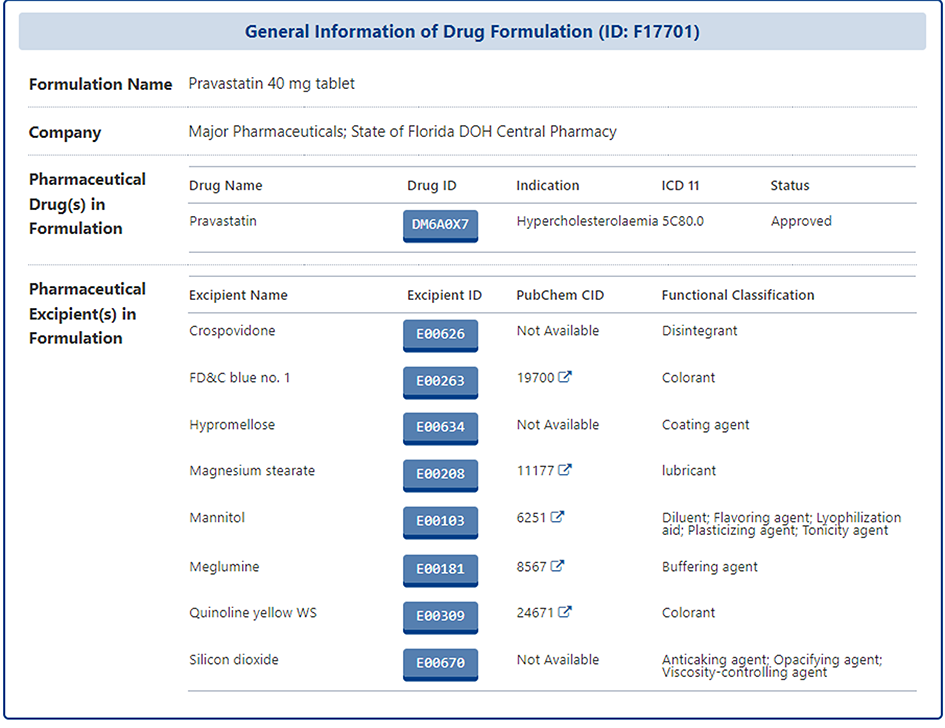
In the detail page of each specific excipient, DrugMAP firstly provides the general information of the pharmaceutical excipient, which includes the "Name", "Synonyms", "Function", "Formula", "SMILES", "InChI", and etc.

Moreover, all of the drugs that co-administrated with this excipient were provided in form of list.
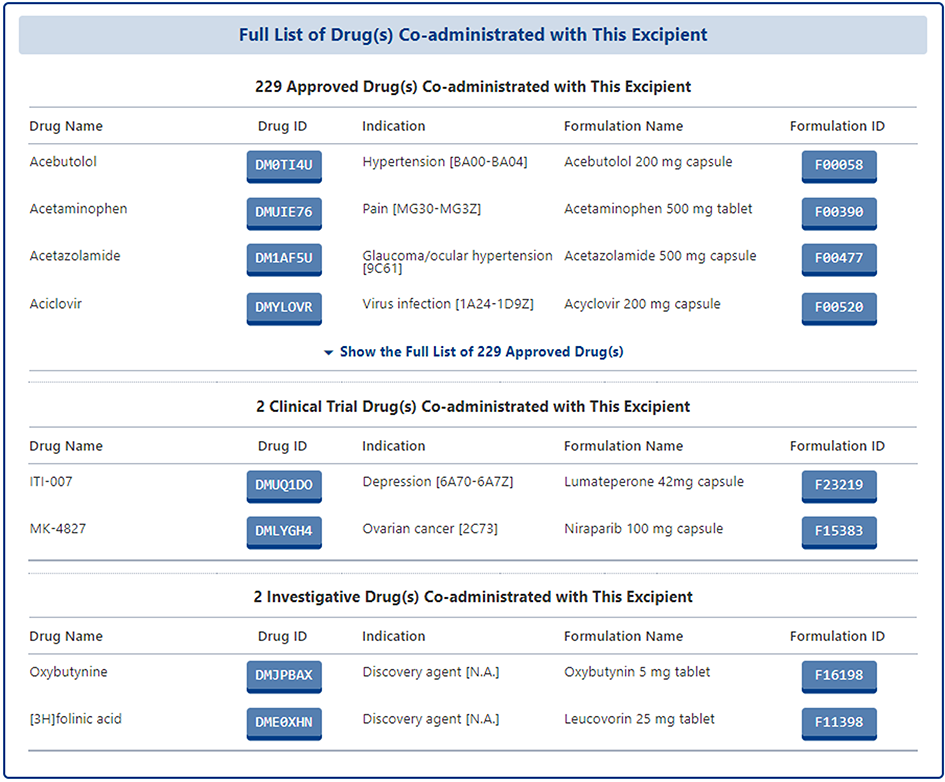
Drug Combination (DrugComb) Profiles of Drug
DrugMAP provides comprehensive information on drug combinations, including not only approved drug combinations in the FDA Orange Book, but also drug combinations that are or have been in clinical studies as documented on the ClinicalTrial.gov website. Secondly, DrugMAP also integrates drug combination high-throughput screening data from multiple sources, normalizes the synergy scores of different drug combinations tested for the same drug, and provides corresponding heatmaps to provide users with a quick overview of the drug combination high-throughput screening results of the drug.
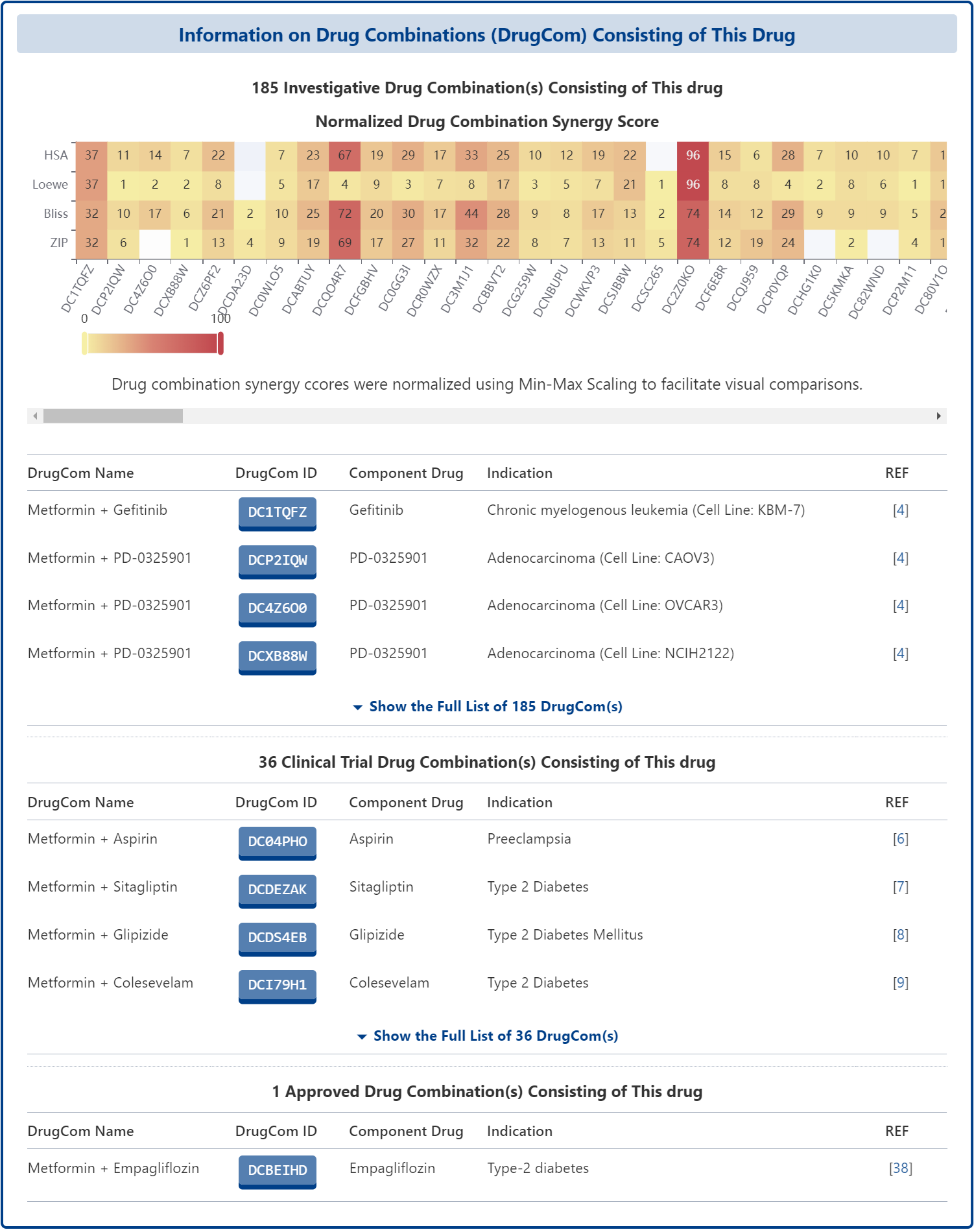
Drug Combination Information in DrugMAP
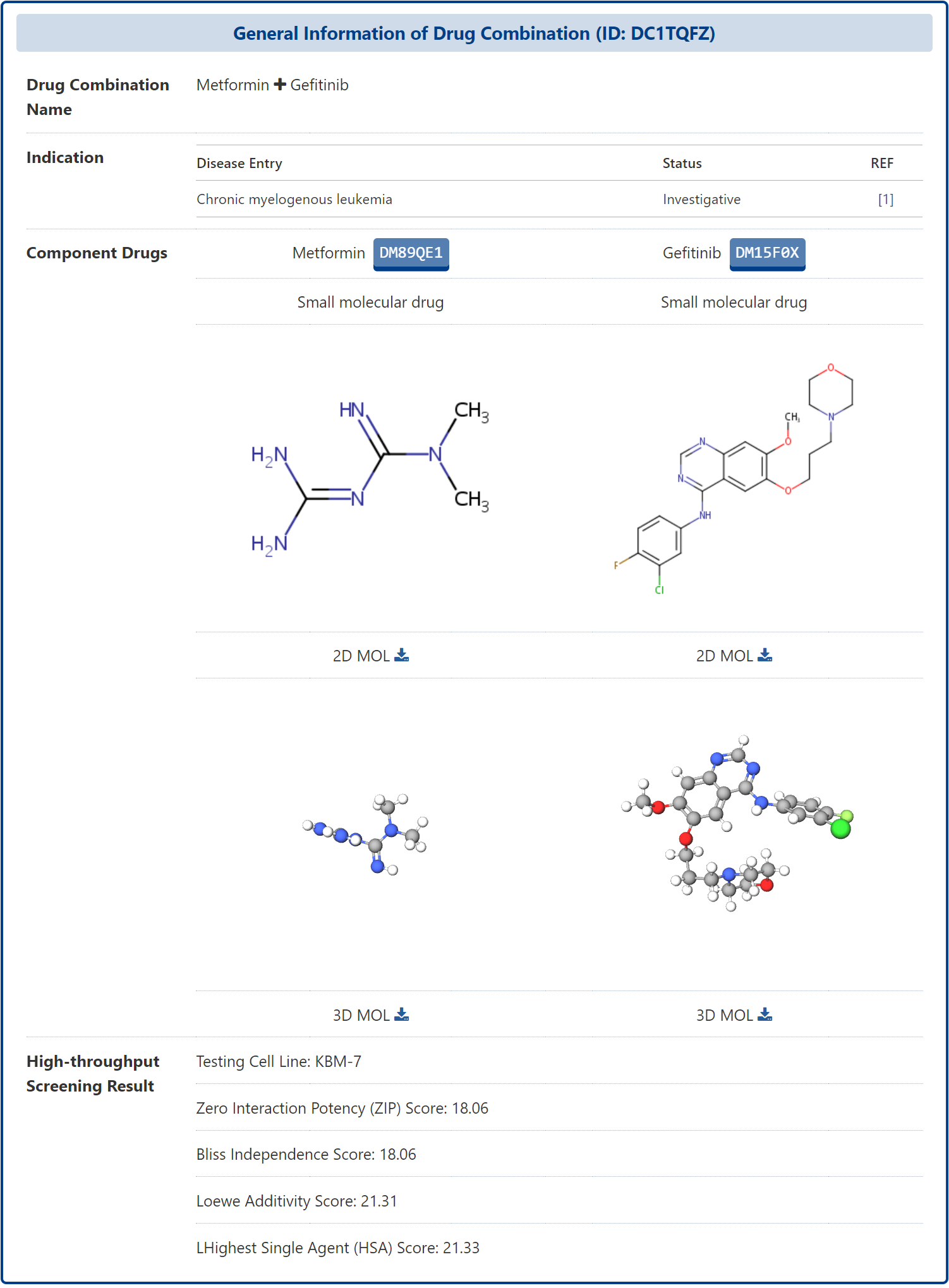
On the Drug Combination (DrugComb) Profiles of Drug page, you can click on the ID of a drug combination to view detailed information about that drug combination, known as Drug Combination Information. DrugMAP firstly provides the basic information of the drug combination and the comparison of the basic information of the different components of the drug combination, including the indication of the drug combination, the name, type and structure of the component drugs, etc.
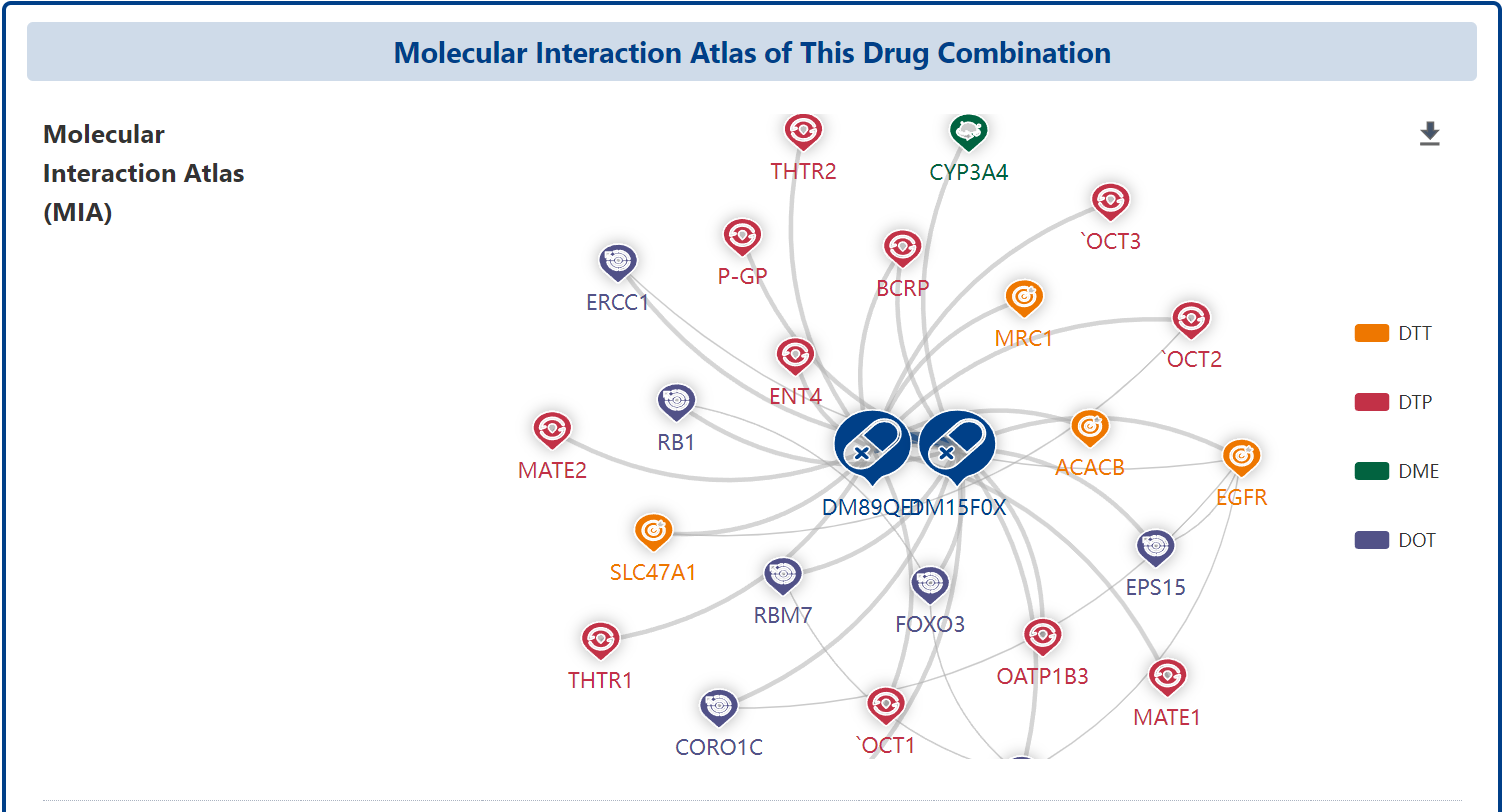
In addition, the page also provides the molecular interaction atlas of this drug combination, which visualizes the different interacting biomolecules of the component drugs of the drug combination in the form of interaction networks to help users understand the mechanism of action of the drug combination in a comprehensive manner.
Drug Reposition (DrugRepo) Profiles of Drug
DrugMAP's “Drug Reposition (DrugRepo) Profiles of Drug” provides comprehensive information on the drug's indications, as well as mapping the drug's interactions with biomolecules in the body across different diseases in the form of interaction networks at the bottom. In addition to providing comprehensive indication information, the “DrugRepo Profiles of Drugs” also maps the drug's interactions with biomolecules in different diseases in the form of interaction networks below, to help users understand the molecular mechanisms of disease repositioning
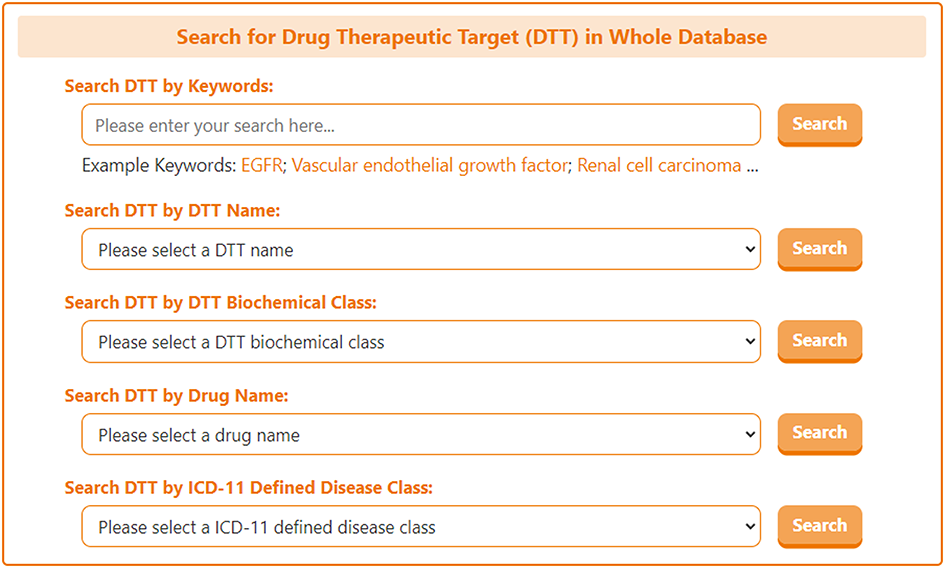
On the search page for DTT, DrugMAP offers a keyword search field for DTT and 4 drop down menu search fields, the 4 drop-down menus provide information about the DTT's "DTT Name", "DTT Biochemical Class", "Drug Name", and "ICD-11 Defined Disease Class", the user can select the item they are interested in to search for drug targets related to this item.
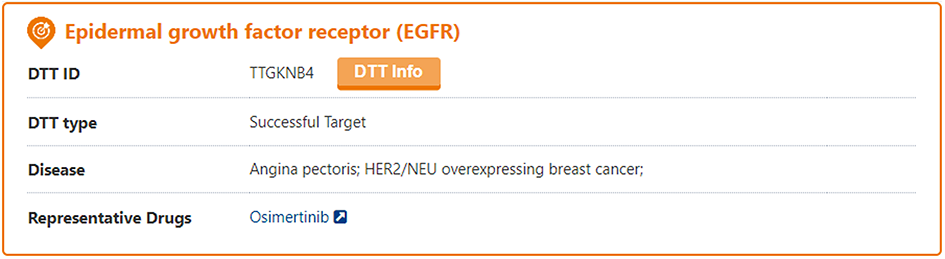
The search results page for a DTT briefly displays information about DTT Name, DTT Type, Disease, and Representative Drug. And users can click the "DTT Info" button to access the detail page of DTT.
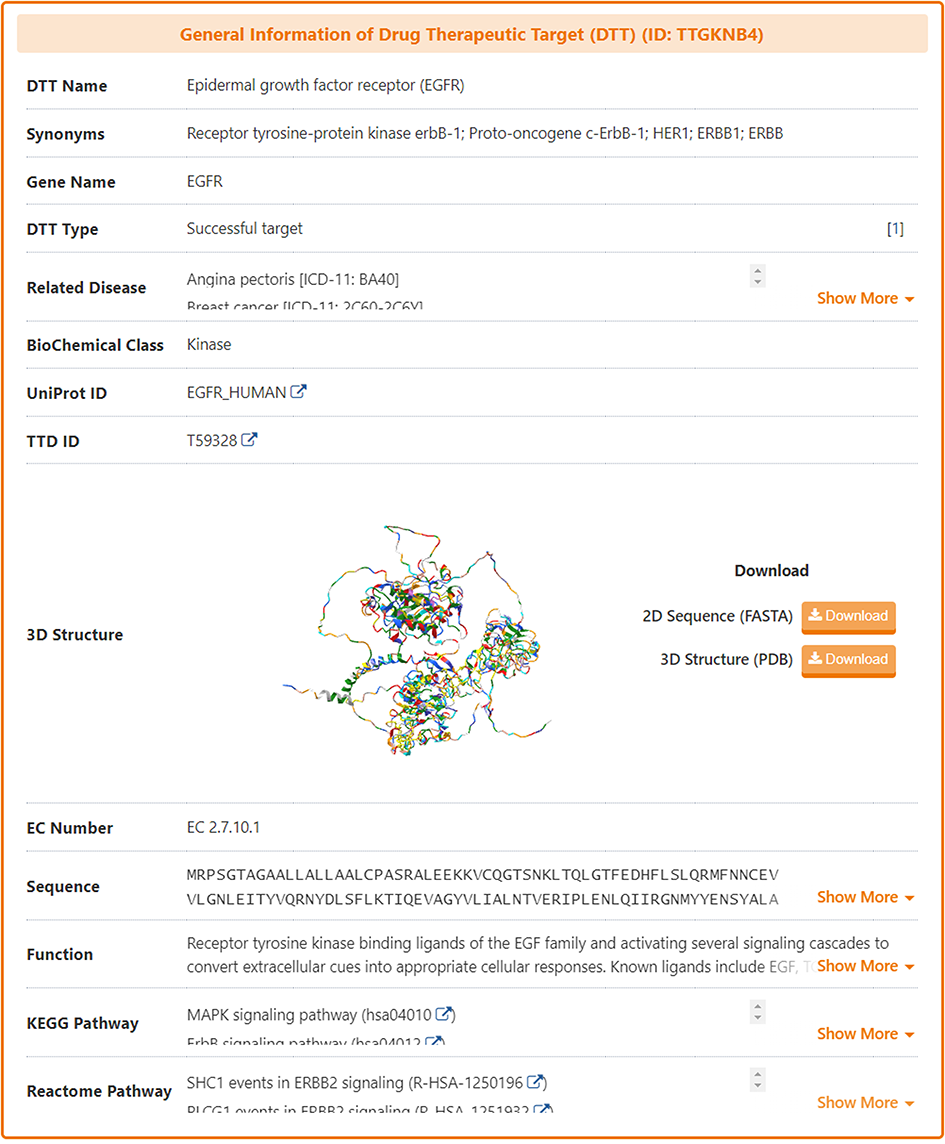
In the "General Information of Drug Therapeutic Target (DTT)" section of the DTT information page, DrugMAP provides information on DTT Name, Synonyms, Gene Name, DTT Type, Related Disease, BioChemical Class, UniProt ID, 3D Structure (visualized via AlphaFold2), EC Number, Sequence, Function, KEGG & Reactome Pathway.
Molecular Interaction Atlas of DTT
In the "Molecular Interaction Atlas of This DTT" section, DrugMAP provides brief information about Drug Name, Indication, Mode of Action (MOA), Highest Status, and reference about all approved, clinical, discontinued, patented, preclinical, and investigative drugs targeting this DTT. Users can jump to the detail information page of drug by clicking the hyperlink under drug name.
Moreover, DrugMAP also provided a molecular interaction map for the subject DTT. The subject DTT is placed in the centre of the map and the drugs that target this DTT are surrounded around it, while the other molecules interacting with these drugs at the periphery.
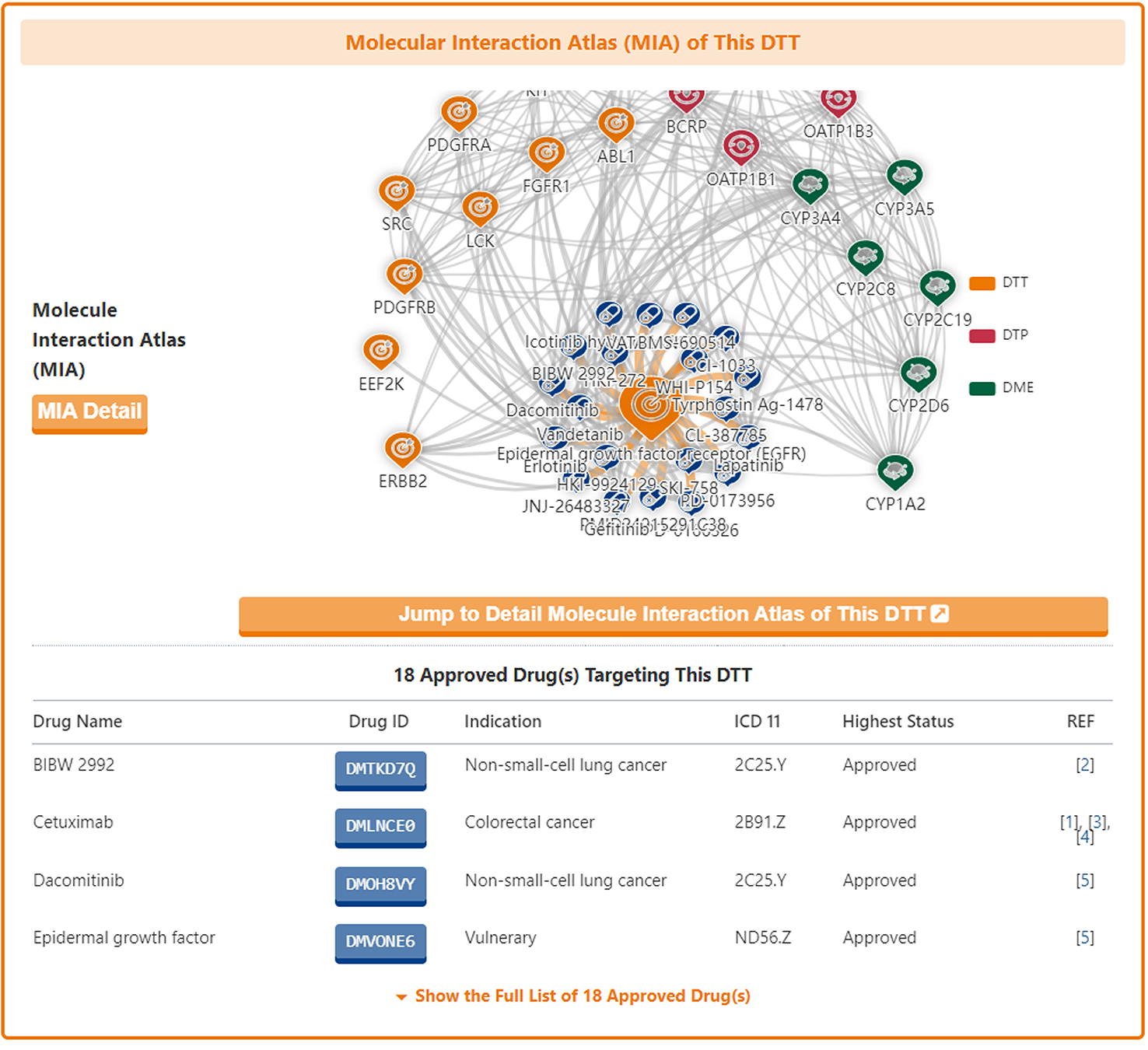
As with the drug information page, a "Jump to Detail Molecular Interaction Atlas of This DTT" button is provided at the bottom of the map to jump to the details page.
Molecular Expression Atlas of DTT
In the "Molecular Expression Atlas of This DTT" section, DrugMAP provides differential expression of subject DTT between healthy and diseased tissues at the site of the lesion under different disease conditions.
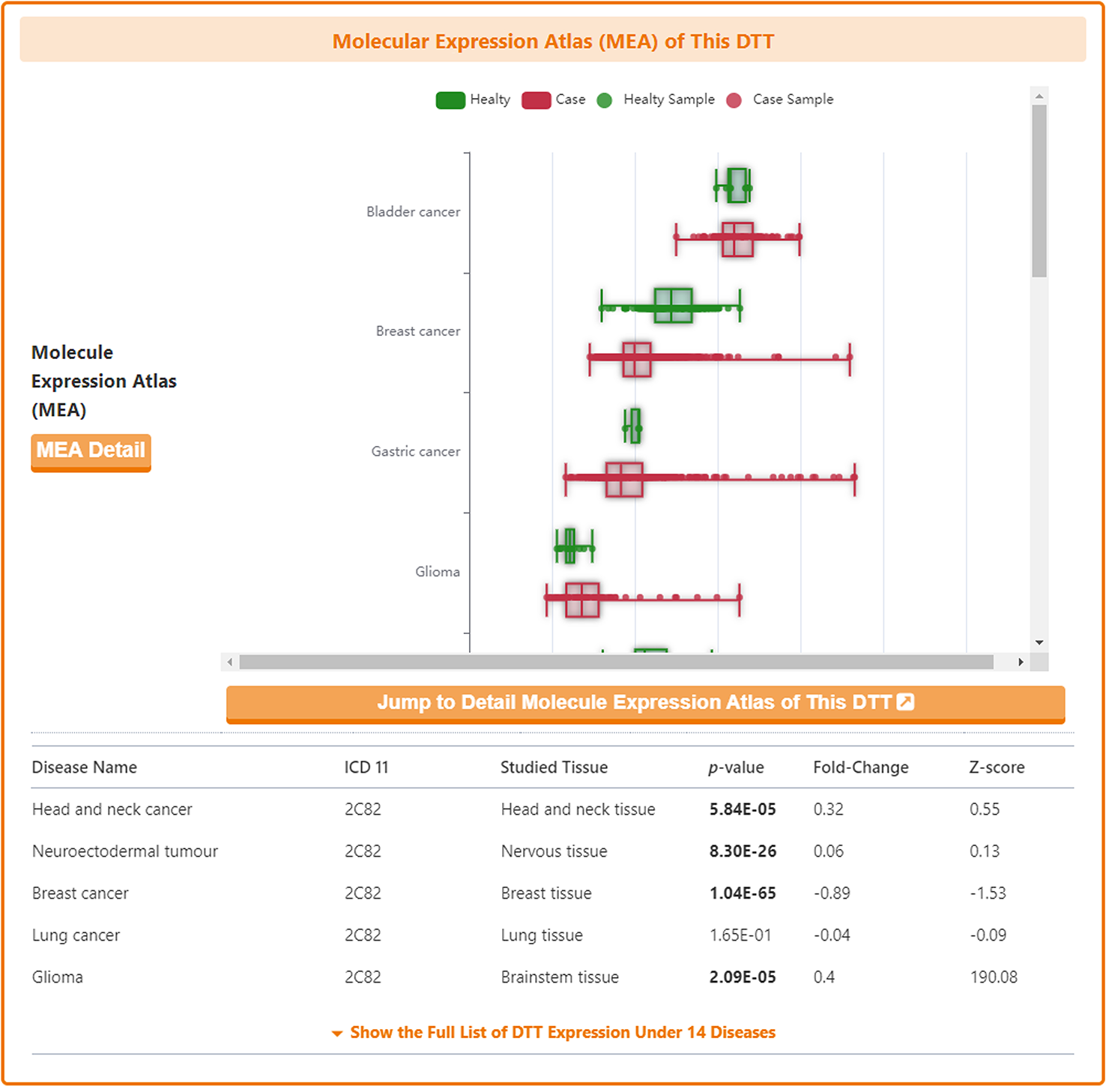
As with the drug information page, a "Jump to Detail Molecular Expression Atlas of This DTT" button is provided at the bottom of the map to jump to the details page.
In the detail page, DrugMAP provides information on the expression of subject DTT in different organs of healthy individuals, where four major ADME-related organs are highlighted in red.
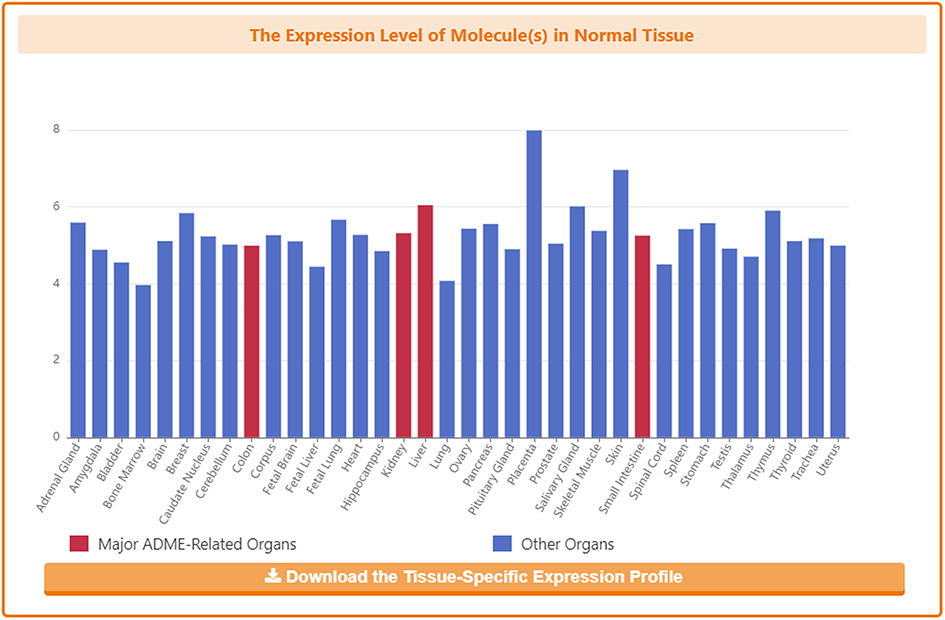
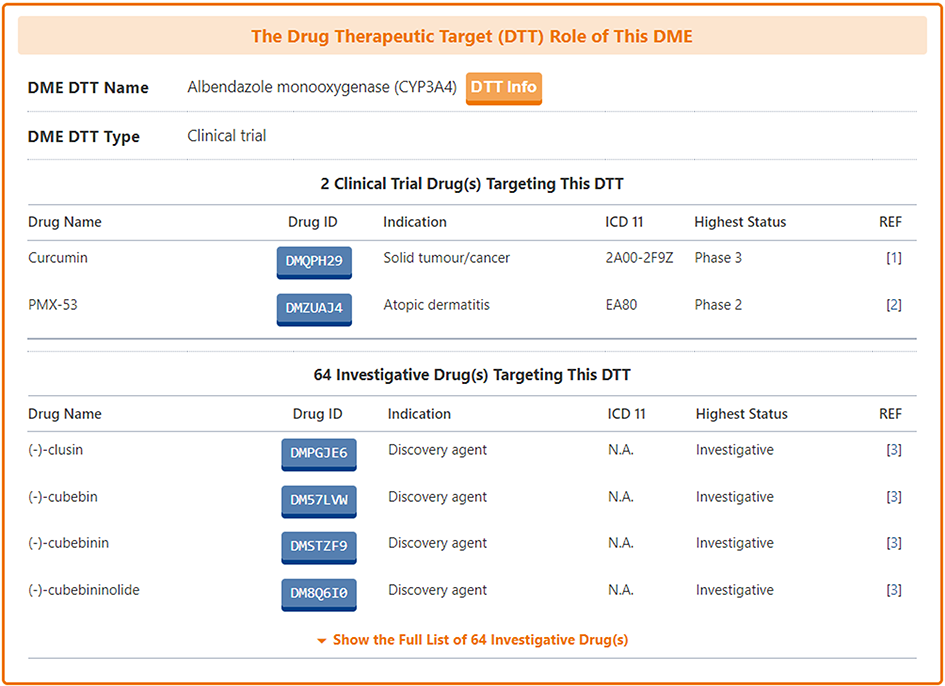
Information on Pharmacokinetic Roles of DTT
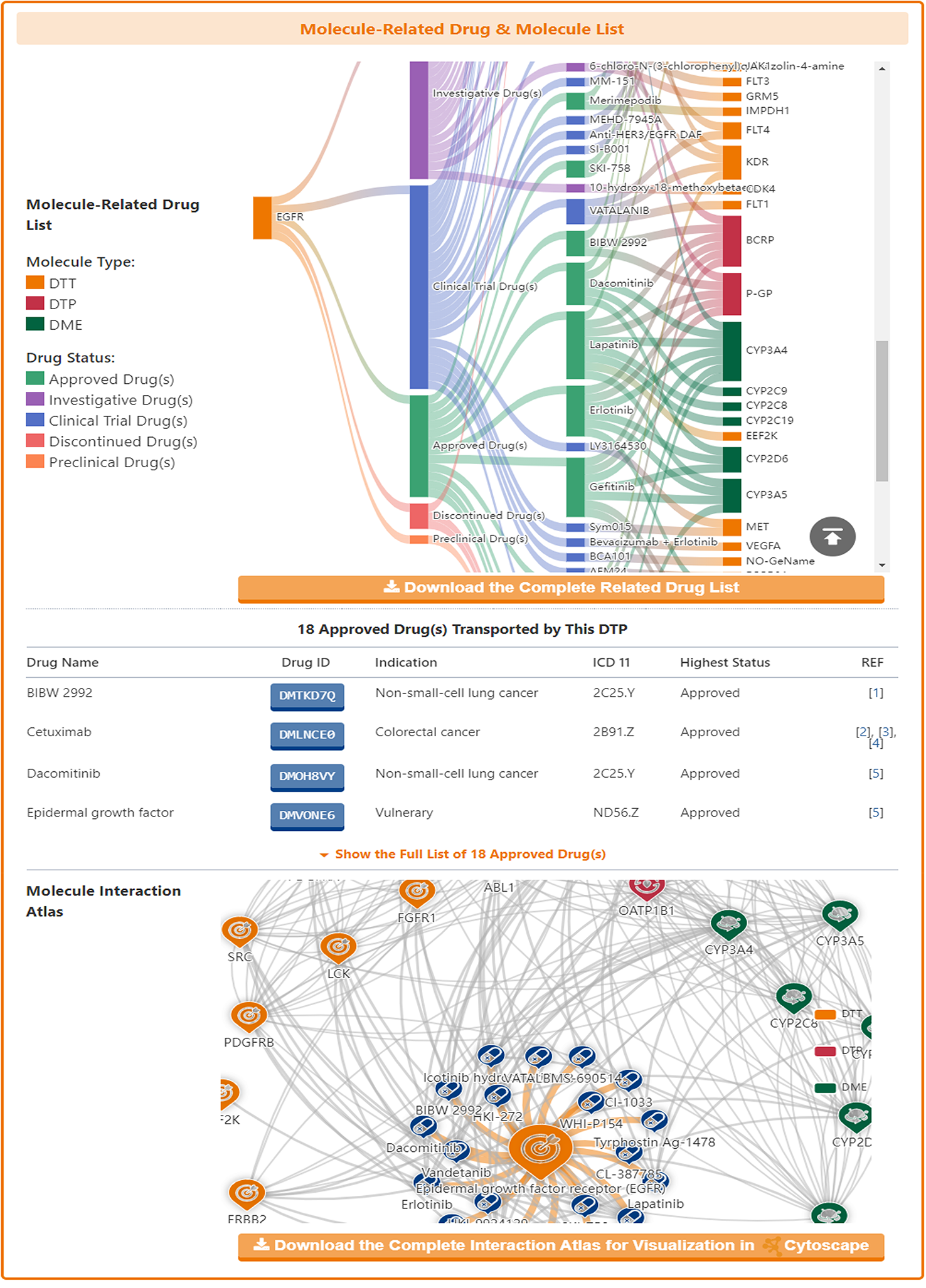
Among the molecules there are some that have multiple identities, acting as drug targets, drug transporters or drug-metabolizing enzymes. DrugMAP pays particular attention to these multiple-roles molecules and provides information on them in the "The Drug Transporter (DTP)/Drug-Metabolizing Enzyme (DME) Role of This Target" section.
On the search page for DTP, DrugMAP offers a keyword search field for DTPs and 4 drop down menu search fields, the 4 drop-down menus related to DT provide information about the DTP's "DTP Name", "DTP Family", "Drug Name", and "ICD-11 Defined Disease Class", the user can select the item they are interested in to search for DTs related to this item.
The search results page for a DTP briefly displays information about DTP Name, DTP Family, Representative Substrate(s), and Representative Drug. And users can click the "DTP Info" button to access the detail page of DTP.
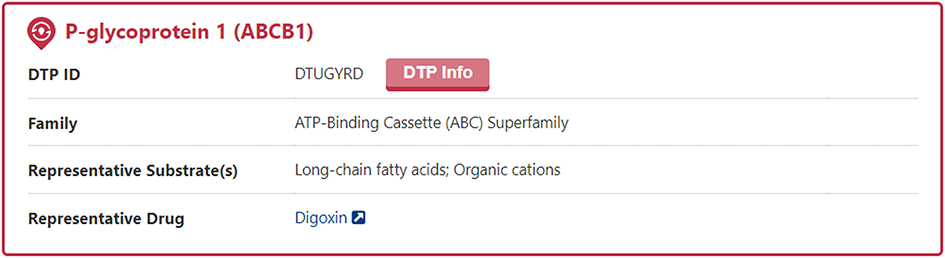
In the "General Information of Drug Transporter (DTP)" section of the DTP information page, DrugMAP provides information on DTP Name, DTP Synonyms, Gene Name, UniProt ID, DT Family, Tissue Specificity, Function, Endogenous Substrate(s), 3D Structure (visualized via AlphaFold2), TCDB ID, Gene ID.
Molecular Interaction Atlas of DTP
In the "Molecular Interaction Atlas of This DTP" section, DrugMAP provides brief information about Drug Name, Indication, Mode of Action (MOA), Highest Status, and reference about all approved, clinical, discontinued, patented, preclinical, and investigative drugs targeting this DTT. Users can jump to the detail information page of drug by clicking the hyperlink under drug name.
Moreover, DrugMAP also provided a molecular interaction map for the subject DTP. The subject DTP is placed in the centre of the map and the drugs that target this DTP are surrounded around it, while the other molecules interacting with these drugs at the periphery.
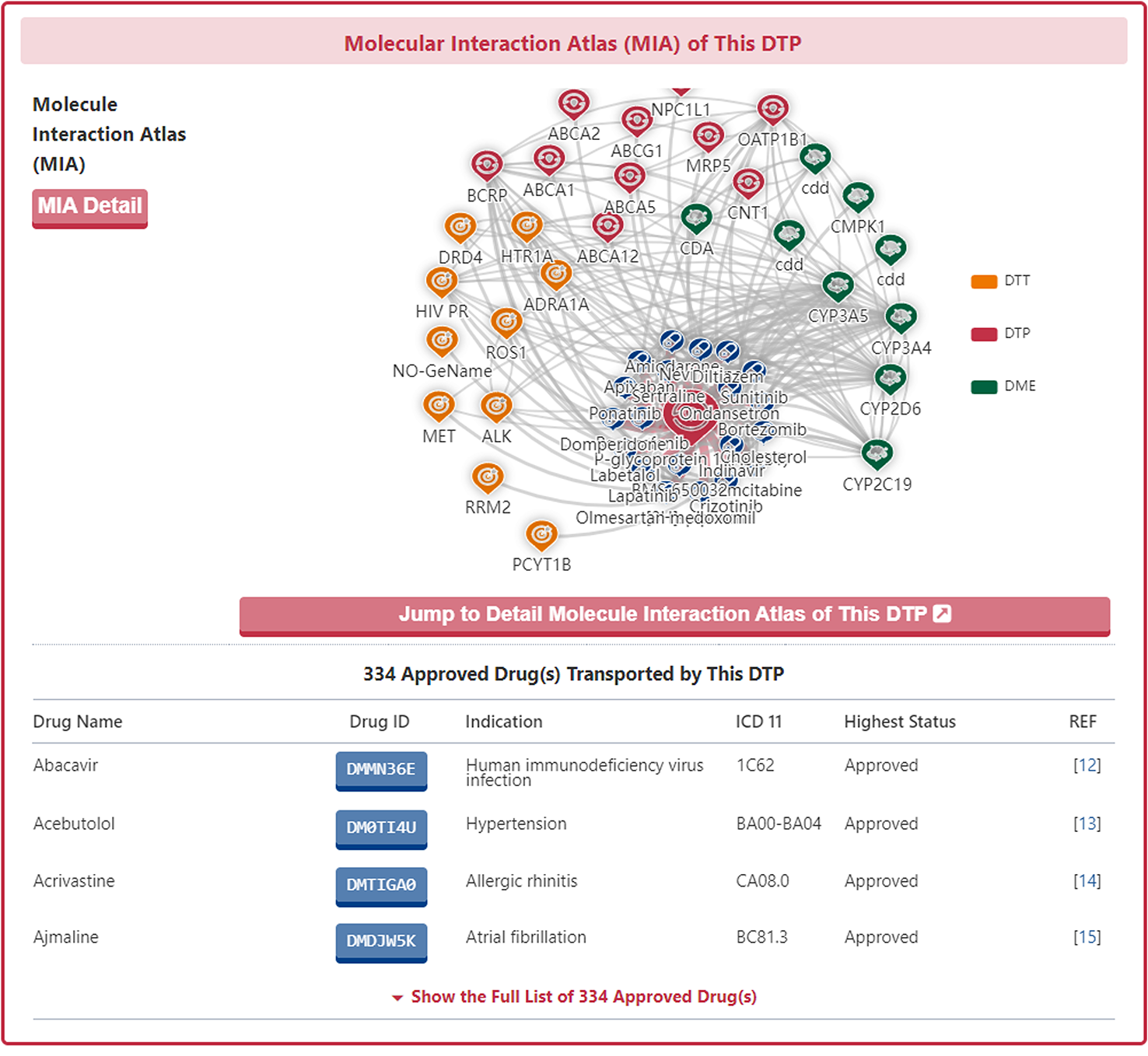
As with the drug information page, a "Jump to Detail Molecular Interaction Atlas of This DTP" button is provided at the bottom of the map to jump to the details page.
Molecular Expression Atlas of DTP
In the "Molecular Expression Atlas of This DTP" section, DrugMAP provides differential expression of subject DTP between healthy and diseased tissues at the site of the lesion under different disease conditions.
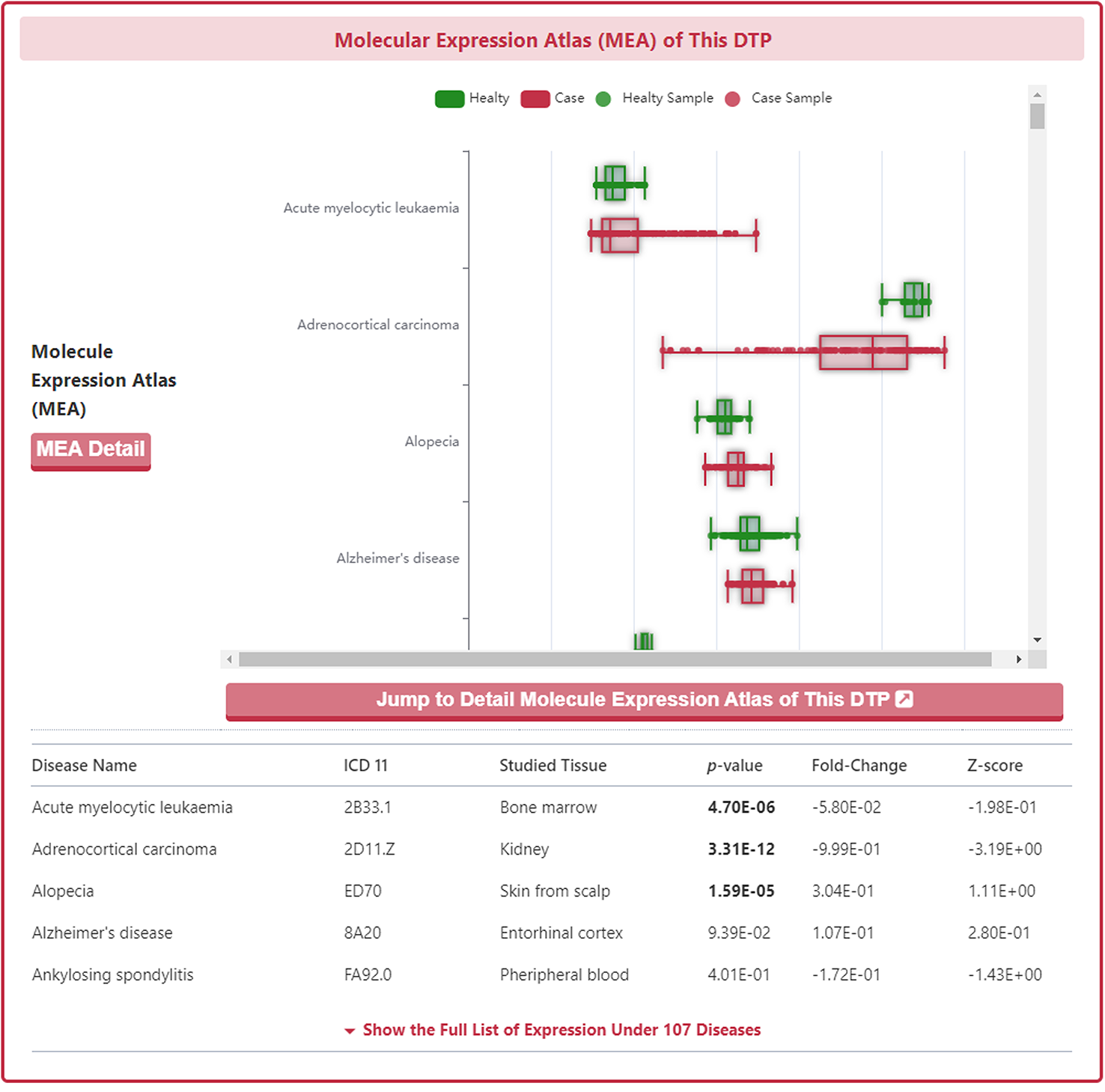
As with the drug information page, a "Jump to Detail Molecular Expression Atlas of This DTP" button is provided at the bottom of the map to jump to the details page.
In the "Drug Affinity of This DT Assessed by Cell Line" section, DrugMAP provides Cell Line and Affinity data about all approved, clinical, discontinued, patented, preclinical, and investigative drugs transported by this DTP. Users can jump to the detail information page of drug by clicking the hyperlink under drug name.
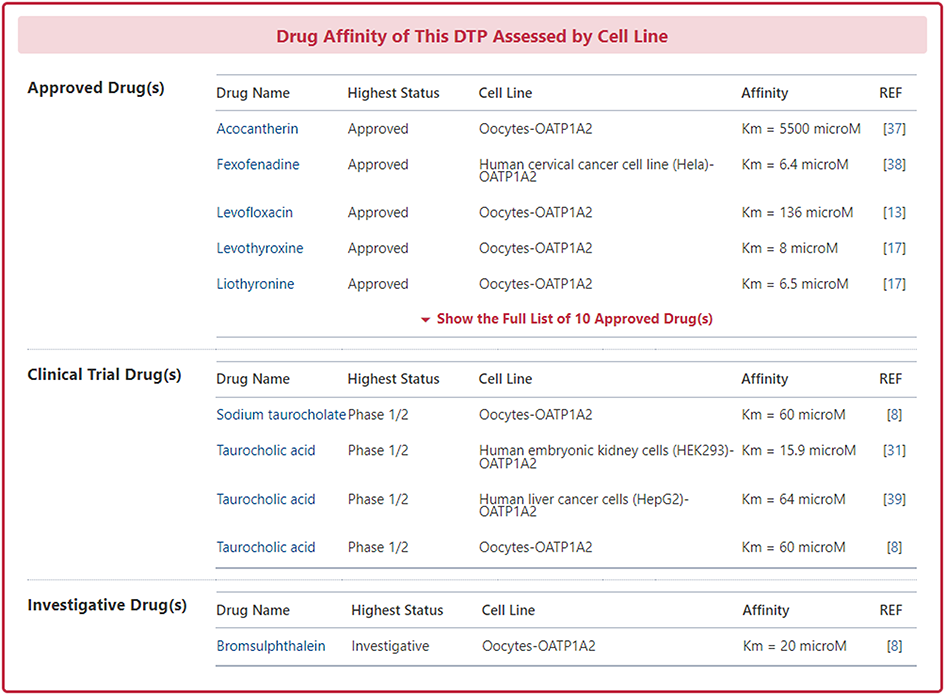
Information on Pharmacodynamic Roles of DTP
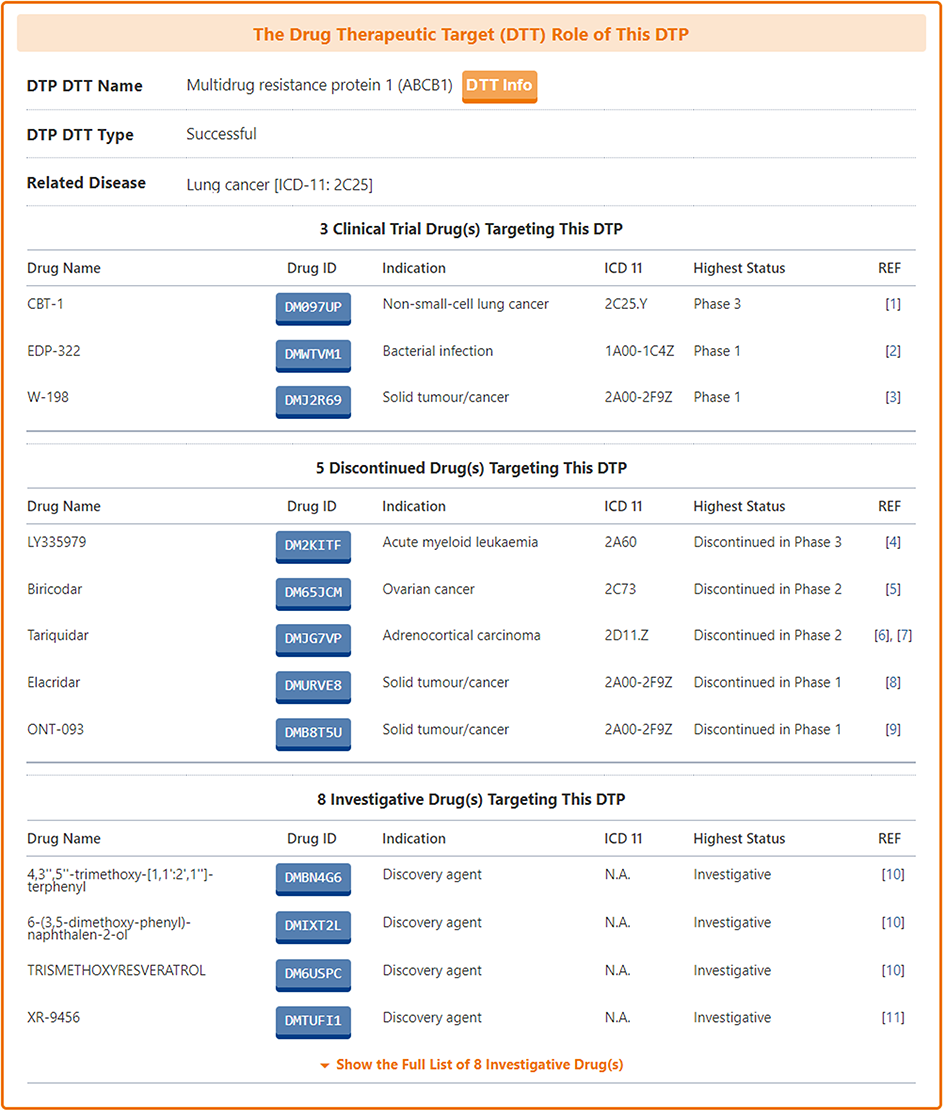
For those DTPs that can be used as drug targets, DrugMAP pays particular attention to provide information on them in the "The Drug Therapeutic Target (DTT) Role of This DTP" section.
On the search page for drug-metabolizing enzyme (DME), DrugMAP offers a keyword search field for DMEs and 4 drop down menu search fields, the 4 drop-down menus related to DT provide information about the DT's "DME Name", "DME Species Name", "Drug Name", and "ICD-11 Defined Disease Class", the user can select the item they are interested in to search for DMEs related to this item.
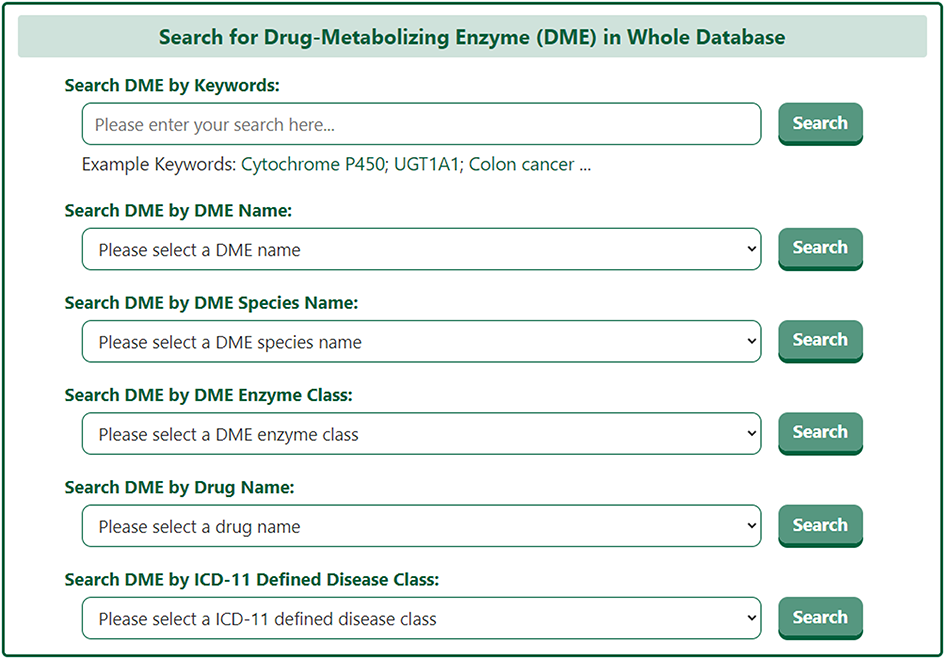
The search results page for a DME briefly displays information about DME Name, EC Number, Tissue Distribution, and Representative Drug.
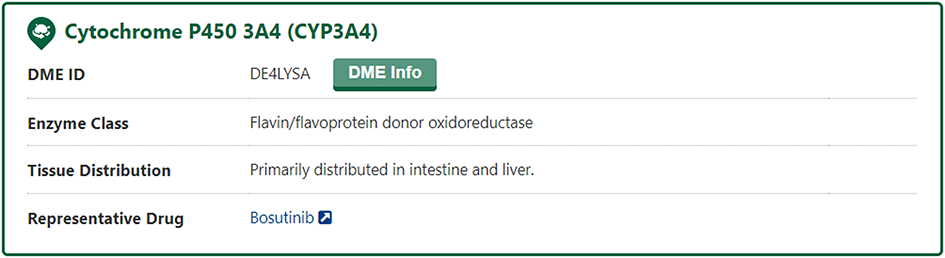
In the "General Information of Drug-Metabolizing Enzyme (DME)" section of the DME information page, DrugMAP provides information on DME Name, DME Synonyms, Gene Name, Gene ID, UniProt ID, EC Number, Lineage, Function, specifically, DrugMAP provided a INTEDE-hyperlink of this DME.
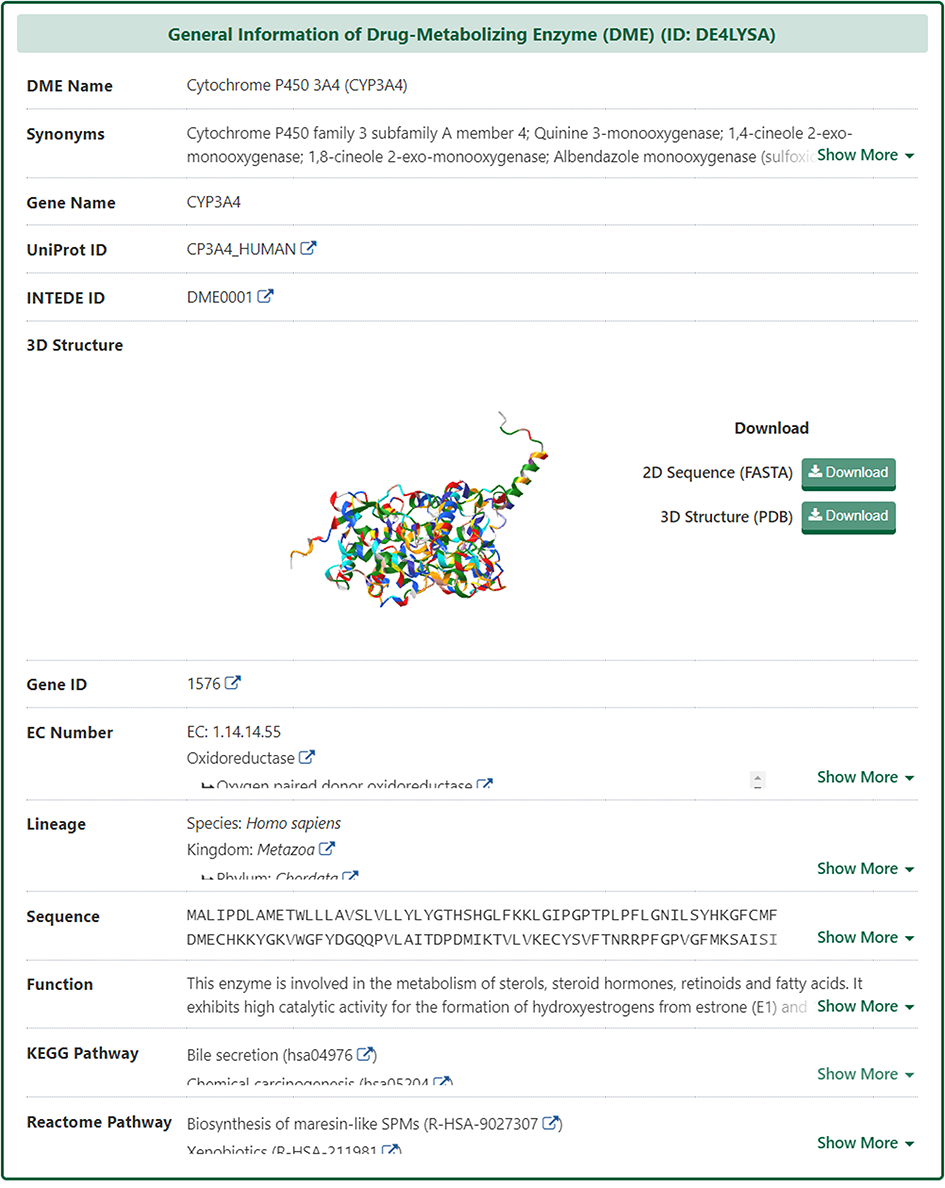
Molecular Interaction Atlas of DME
In the "Molecular Interaction Atlas of This DME" section, DrugMAP provides brief information about Drug Name, Indication, Mode of Action (MOA), Highest Status, and reference about all approved, clinical, discontinued, patented, preclinical, and investigative drugs targeting this DME. Users can jump to the detail information page of drug by clicking the hyperlink under drug name.
Moreover, DrugMAP also provided a molecular interaction map for the subject DME. The subject DME is placed in the centre of the map and the drugs that target this DME are surrounded around it, while the other molecules interacting with these drugs at the periphery.
As with the drug information page, a "Jump to Detail Molecular Interaction Atlas of This DME" button is provided at the bottom of the map to jump to the details page.
Molecular Expression Atlas of DME
In the "Molecular Expression Atlas of This DME" section, DrugMAP provides differential expression of subject DME between healthy and diseased tissues at the site of the lesion under different disease conditions.
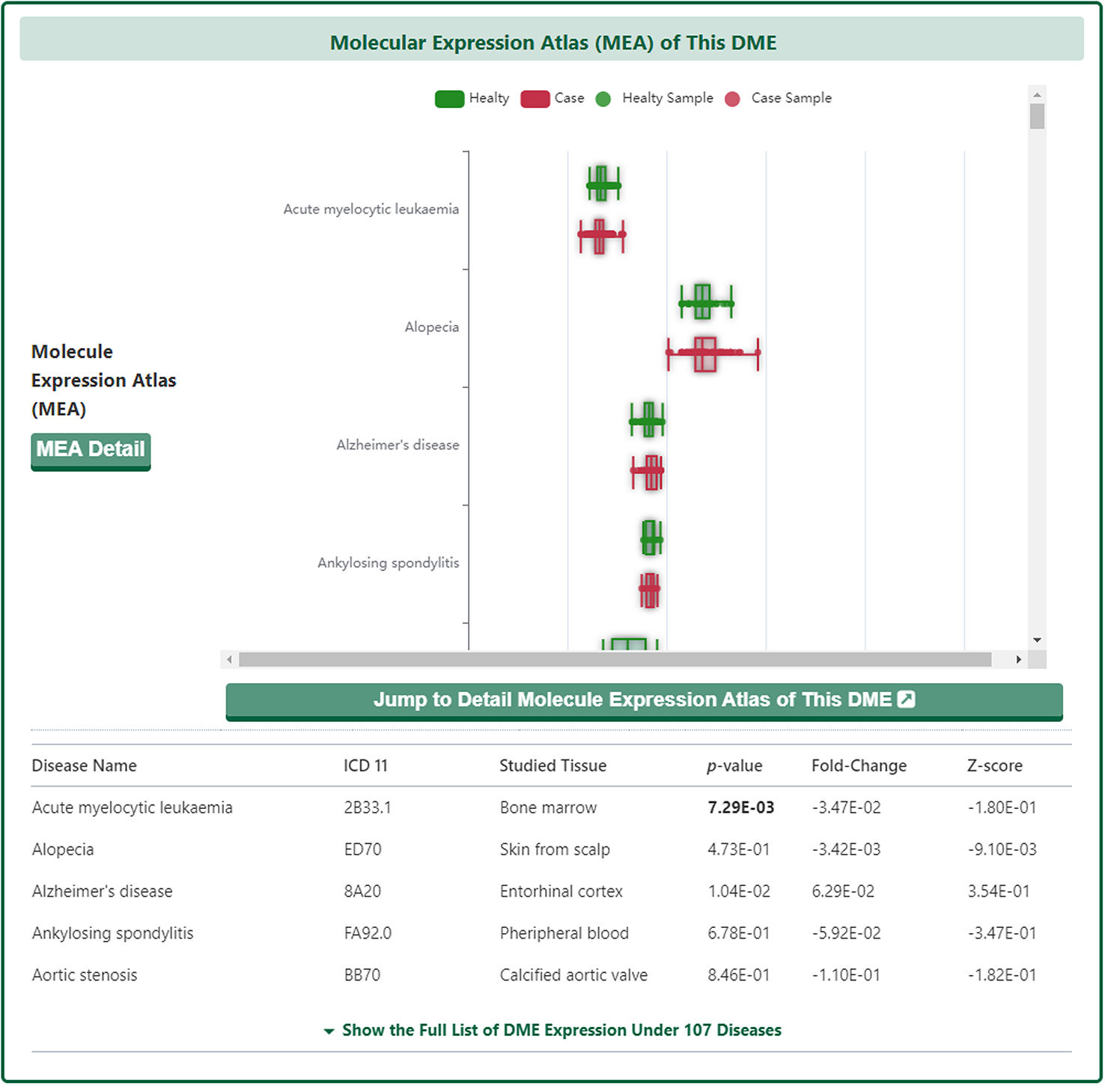
As with the drug information page, a "Jump to Detail Molecular Expression Atlas of This DME" button is provided at the bottom of the map to jump to the details page.
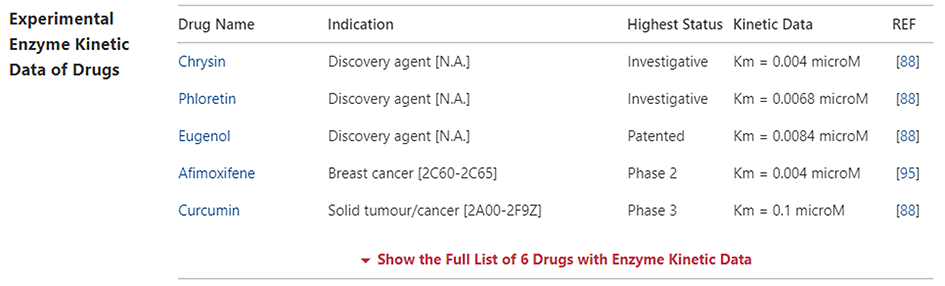
Experimental DME Kinetic Data of Drug
In the "Experimental DME Kinetic Data of Drug" section, DrugMAP provides Cell Line and Affinity data about all approved, clinical, discontinued, patented, preclinical, and investigative drugs transported by this DME. Users can jump to the detail information page of drug by clicking the hyperlink under drug name.
Information on Pharmacodynamic Roles of DME
For those DMEs that can be used as drug targets, DrugMAP pays particular attention to provide information on them in the "The Drug Therapeutic Target (DTT) Role of This DME" section.
On the drug-off-target (DOT) search page, DrugMAP provides a DOT keyword search field and 4 drop-down menu search fields. The 4 drop-down menus related to DOT provide information such as “DOT Name”, “DOT Protein Family", ‘Drug Name’, and ‘ICD-11 Defined Disease Category’ information for the DOT, which allows the user to select the item of interest and search for DMEs related to that item.
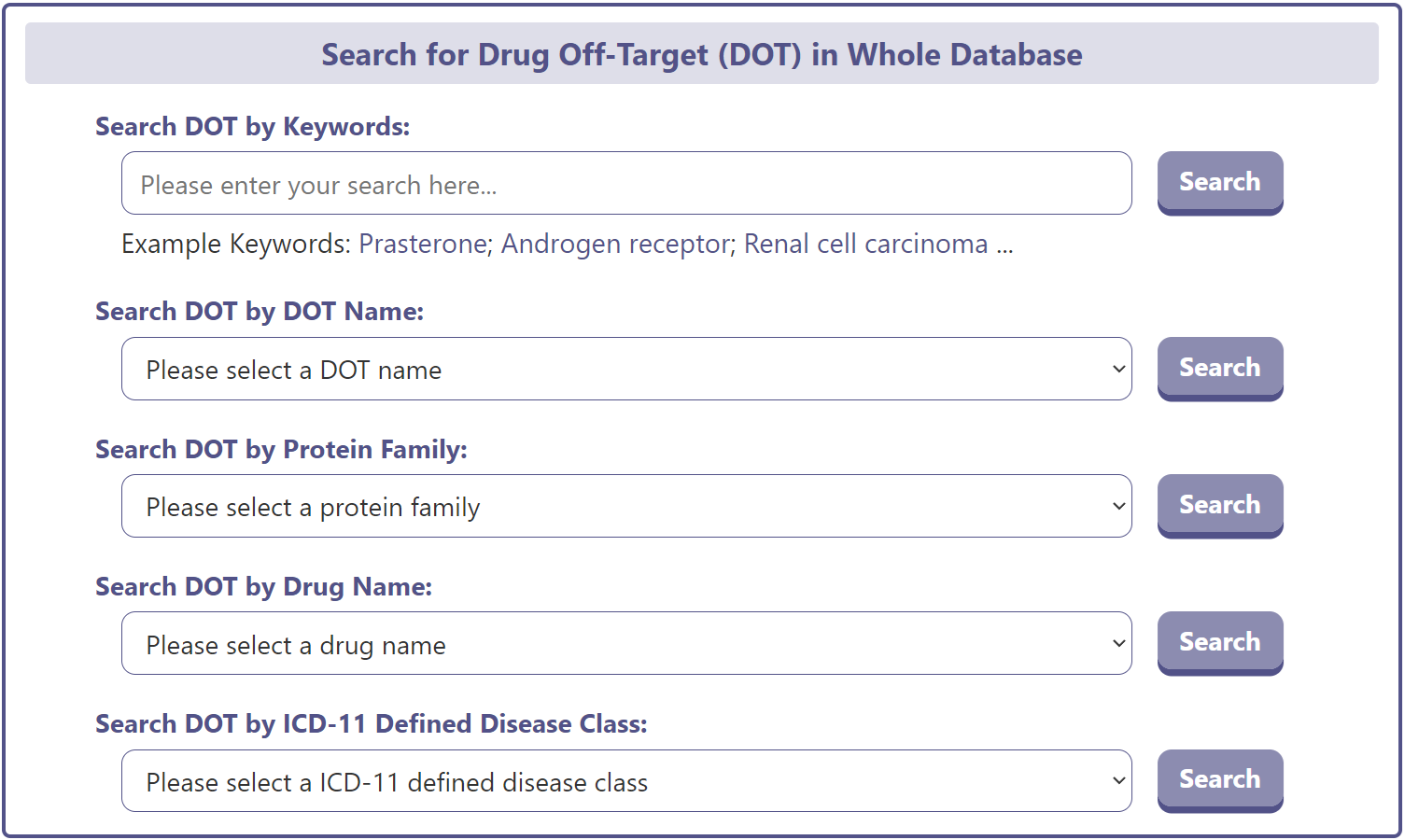
The DOT search results page displays brief information about the DOT name, UniProt ID, related diseases, and representative drugs.
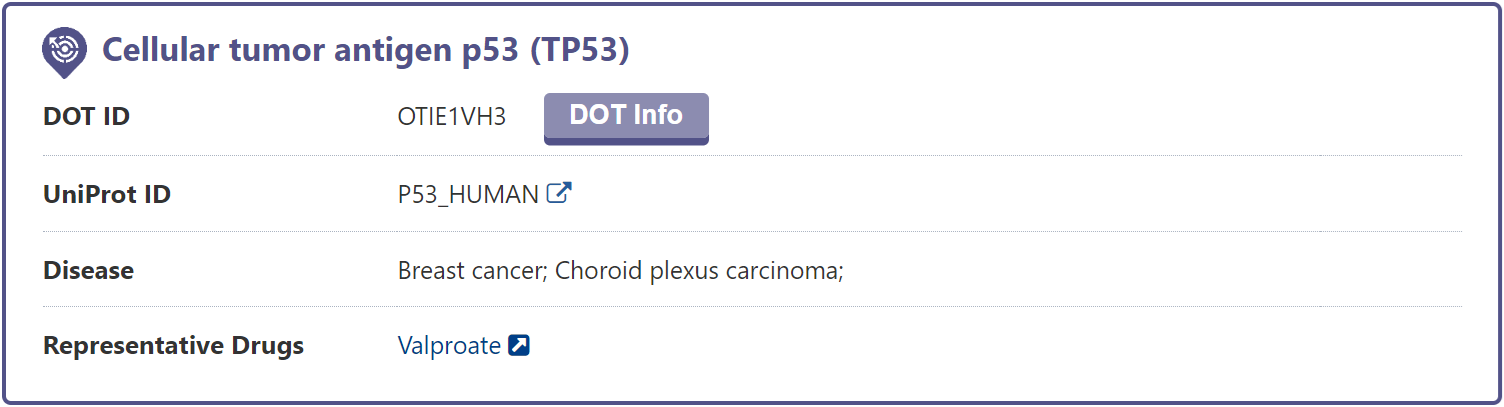
In the "General Information of DOT" section of the DME information page, DrugMAP provides information on DOT Name, DOT Synonyms, Gene Name, Gene ID, UniProt ID, EC Number, Lineage, Function.
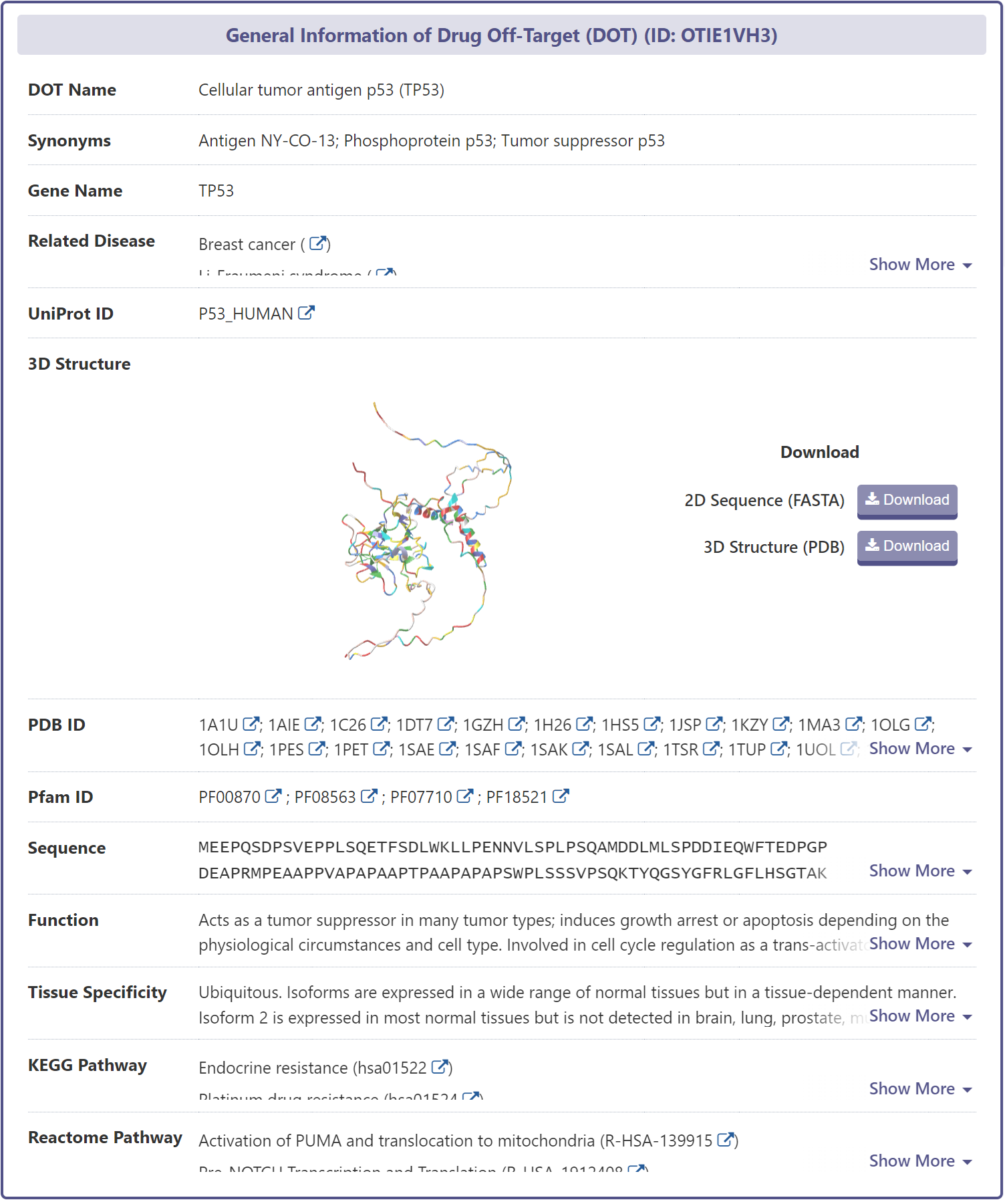
Related Disease Information of DOT
DrugMAP provides a wealth of information on DOT-disease associations from the scientific literature, each of which is accompanied by a corresponding reference.
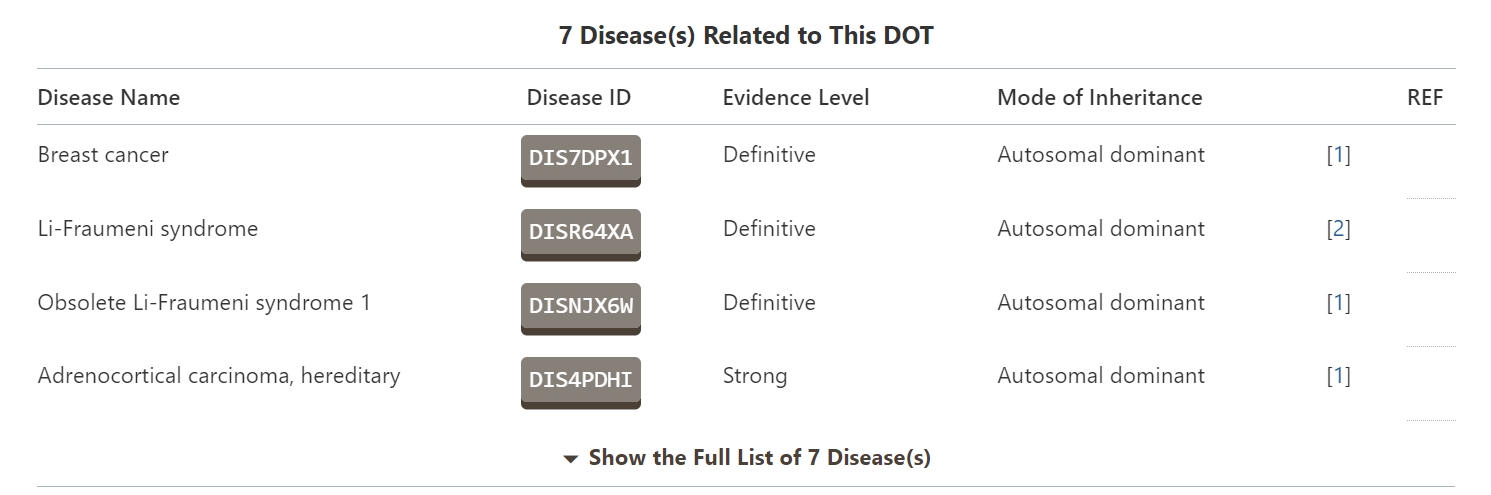
Molecular Interaction Atlas of DOT
In the “Molecular Interaction Atlas of DOT” section, DrugMAP provides brief information about the drug name, indication, mode of action (MOA), highest status, and references to all approved, clinical, discontinued, patented, preclinical, and investigational drugs against this DOT information. Users can jump to the drug details page by clicking on the hyperlink under the drug name.
On the Disease/Drug Indication search page, DrugMAP provides a keyword search field for Disease and 6 drop-down menu search fields that include Drug Name, ICD-11 Defined Disease Class, DTT Name, DTP Name, DME Name, DOT Name, users can select the item of interest and search for Disease associated with that item.
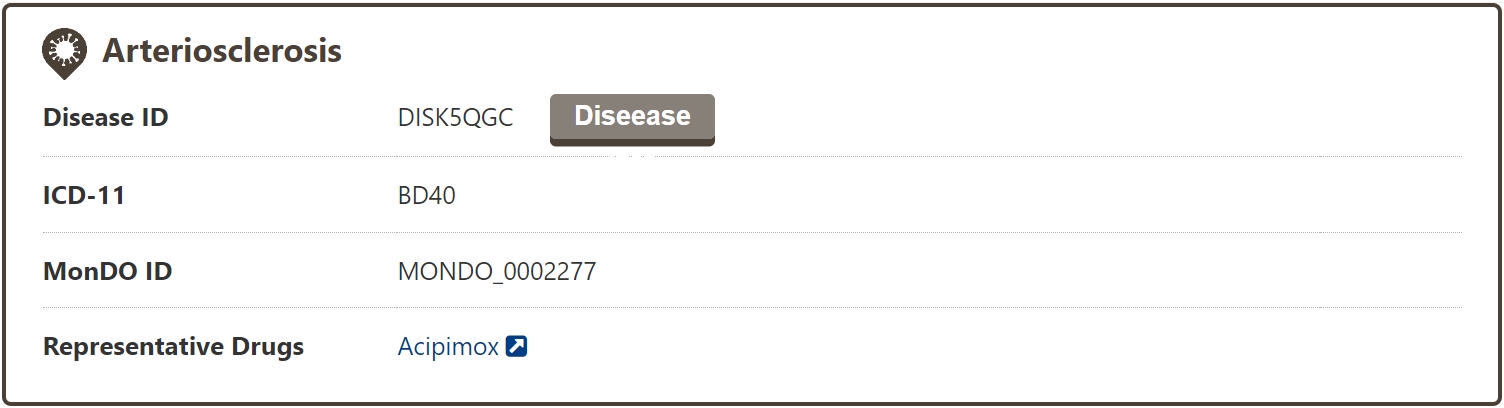
The search results page for a disease shows brief information about the disease name, ICD-11 code, MONDO ID, and representative drugs.
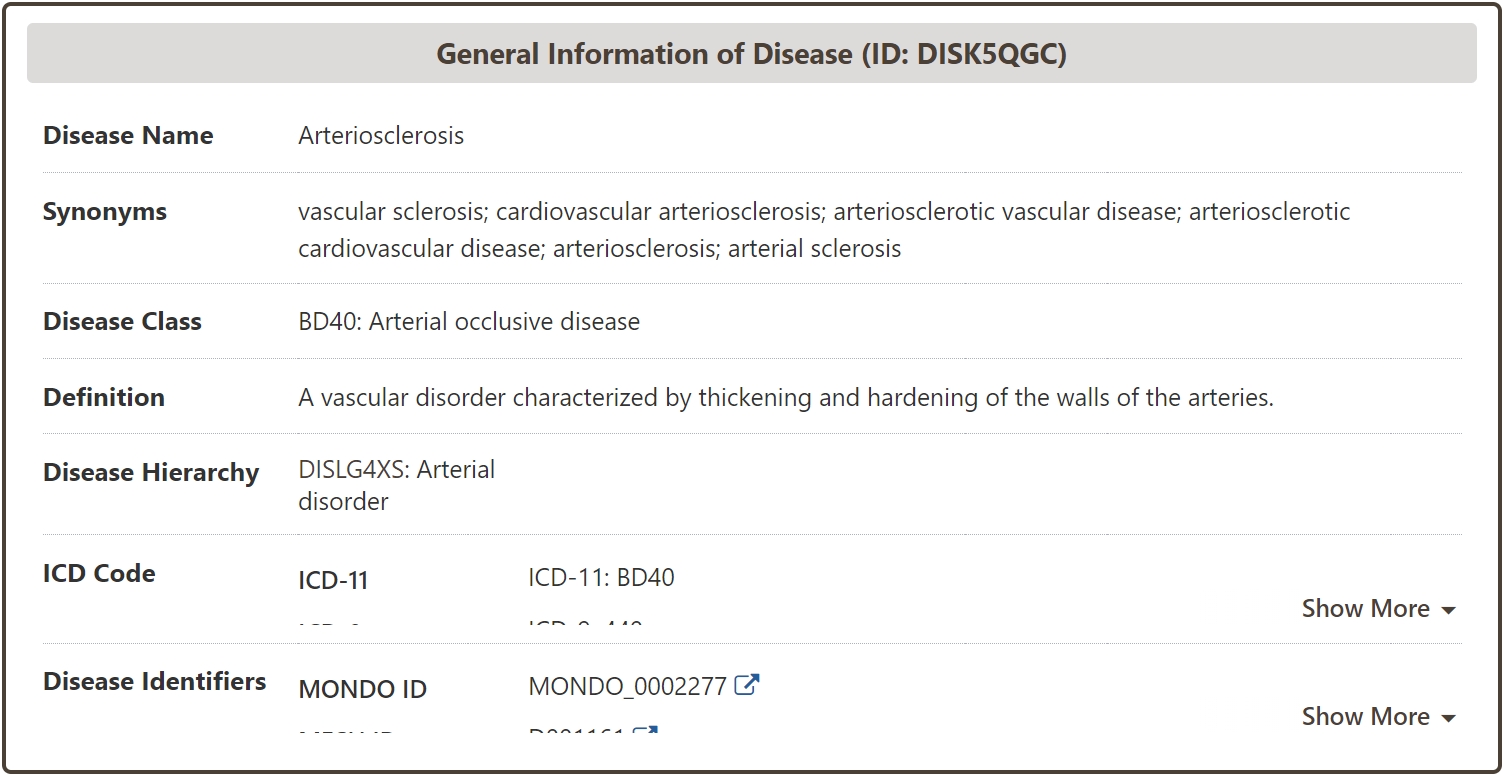
General Information of Disease
In the "General Information of Disease" section of the Disease information page, Information such as Name, Synonym, Class, Definition of the disease is systematically provided in DrugMAP, and Hierachy information of the disease is also provided based on the MONDO ID of the disease. In addition, in order to facilitate data management and data association, DrugMAP also systematically matches and associates diseases with famous disease terminology databases such as ICD and MONDO.
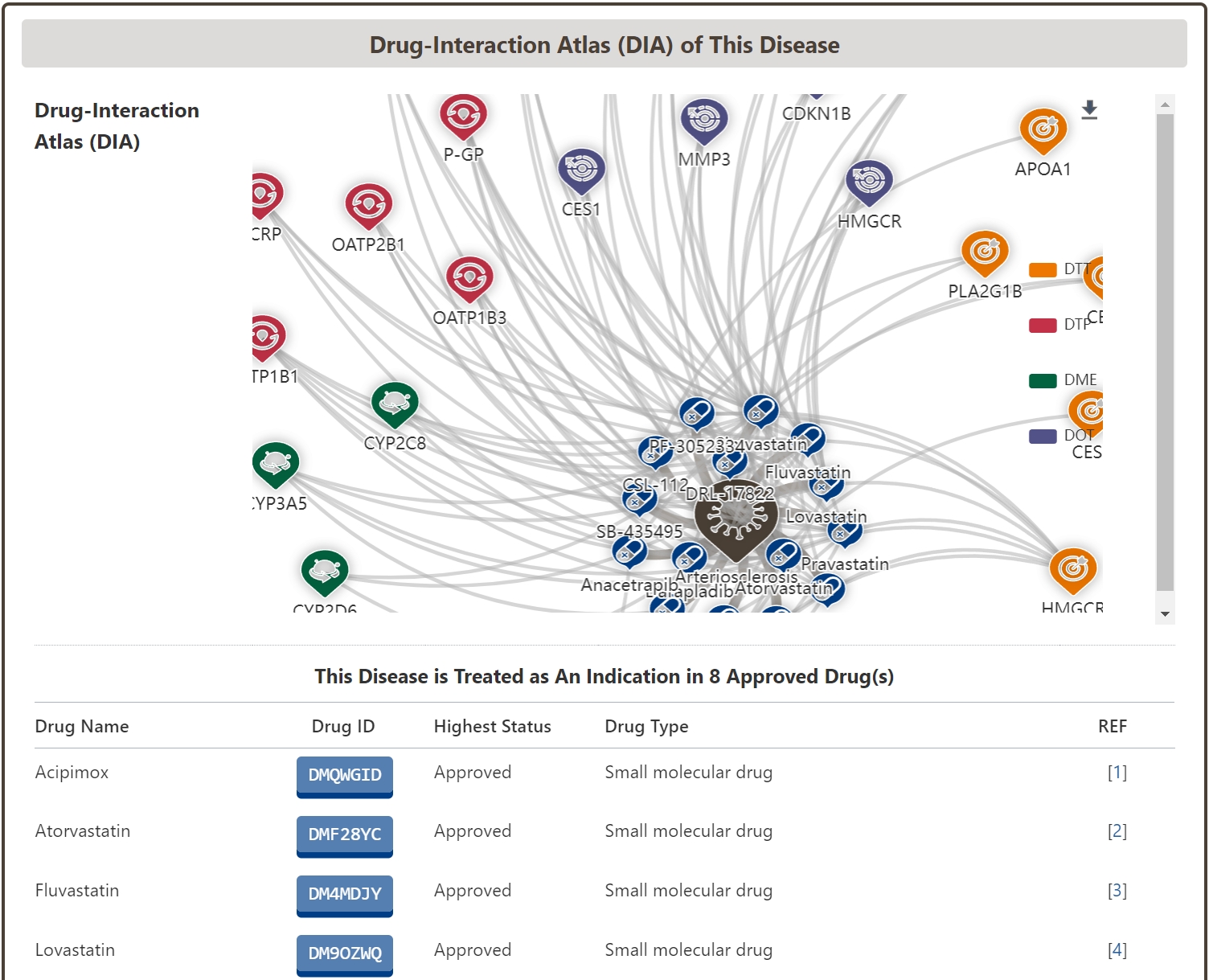
Drug-Interaction Atlas (DIA) of Disease
In the “Drug-Interaction Atlas (DIA) of Disease” section of the Disease Information page, we systematically organize all drugs indicated for the disease, including approved, clinically investigational, and investigational drugs, based on the interacting biological and molecular factors. We also mapped the drug-disease interaction network of the disease based on the interacting biomolecules.
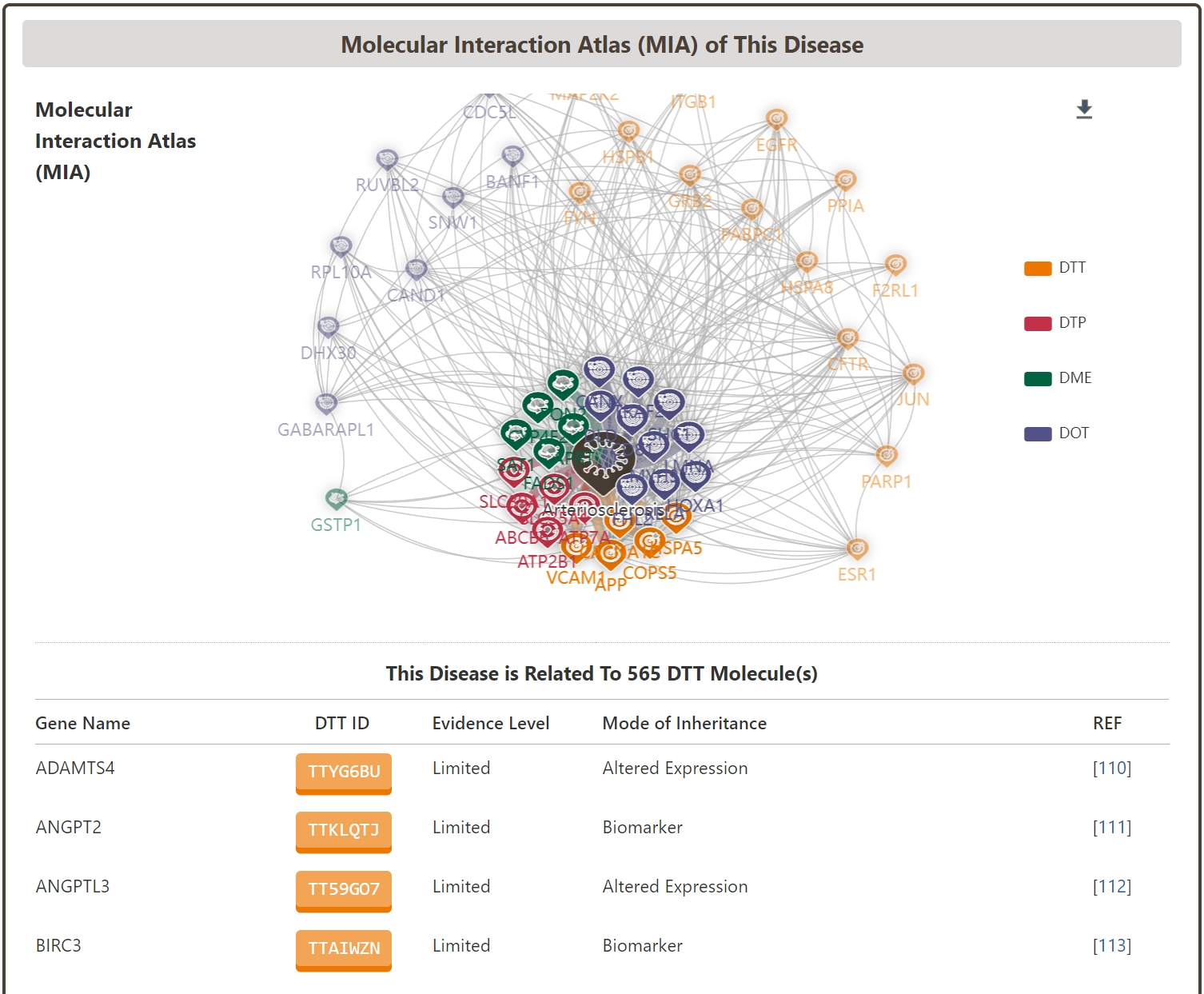
Molecular Interaction Atlas (MIA) of Disease
In addition, the “Molecular Interaction Atlas (MIA) of Disease” section of the Disease Information page systematically provides all the biomolecules associated with the disease, including not only DTT, DTP, and DTP, which are directly related to the pharmacodynamics of the drug, but also DTP, DTP, and DTP, which are directly related to the pharmacodynamics of the drug. These include not only DTT, DTP and DME molecules, which are directly related to drug pharmacodynamics, but also DOT molecules, which are closely related to drug safety and side effects. Through this disease-biomolecule interaction network, users can quickly understand the current status of the disease's biological signaling network, and help to carry out related drug combination and drug repositioning studies.