| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
Citalopram FDA Label
|
| 3 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7547).
|
| 4 |
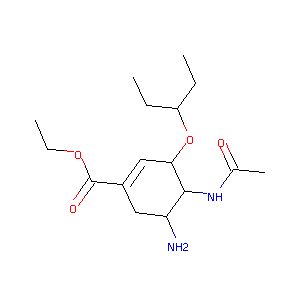
Oseltamivir FDA Label
|
| 5 |
Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007 Mar;70(3):461-77.
|
| 6 |
ClinicalTrials.gov (NCT04261270) A Randomized,Open,Controlled Clinical Study to Evaluate the Efficacy of ASC09F and Ritonavir for 2019-nCoV Pneumonia
|
| 7 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services.
|
| 8 |
MRP1 polymorphisms associated with citalopram response in patients with major depression. J Clin Psychopharmacol. 2010 Apr;30(2):116-25.
|
| 9 |
ABCB1 gene polymorphisms are associated with fatal intoxications involving venlafaxine but not citalopram. Int J Legal Med. 2013 May;127(3):579-86.
|
| 10 |
Organic cation transporters and their pharmacokinetic and pharmacodynamic consequences. Drug Metab Pharmacokinet. 2008;23(4):243-53.
|
| 11 |
PharmGKB summary: citalopram pharmacokinetics pathway. Pharmacogenet Genomics. 2011 Nov;21(11):769-72.
|
| 12 |
Pharmacokinetic-pharmacodynamic relationship of the selective serotonin reuptake inhibitors. Clin Pharmacokinet. 1996 Dec;31(6):444-69.
|
| 13 |
Antidepressant drugs in the elderly--role of the cytochrome P450 2D6. World J Biol Psychiatry. 2003 Apr;4(2):74-80.
|
| 14 |
Citalopram and desmethylcitalopram in vitro: human cytochromes mediating transformation, and cytochrome inhibitory effects. Biol Psychiatry. 1999 Sep 15;46(6):839-49.
|
| 15 |
Identification of three cytochrome P450 isozymes involved in N-demethylation of citalopram enantiomers in human liver microsomes. Pharmacogenetics. 1997 Feb;7(1):1-10.
|
| 16 |
Pharmacokinetics of citalopram in relation to genetic polymorphism of CYP2C19. Drug Metab Dispos. 2003 Oct;31(10):1255-9. doi: 10.1124/dmd.31.10.1255.
|
| 17 |
ABCB1 (MDR1) gene polymorphisms are associated with the clinical response to paroxetine in patients with major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2008 Feb 15;32(2):398-404. doi: 10.1016/j.pnpbp.2007.09.003. Epub 2007 Sep 15.
|
| 18 |
Genetic and clinical predictors of sexual dysfunction in citalopram-treated depressed patients. Neuropsychopharmacology. 2009 Jun;34(7):1819-28. doi: 10.1038/npp.2009.4. Epub 2009 Mar 18.
|
| 19 |
Antioxidant enzyme and malondialdehyde values in social phobia before and after citalopram treatment. Eur Arch Psychiatry Clin Neurosci. 2004 Aug;254(4):231-5.
|
| 20 |
In vitro detection of drug-induced phospholipidosis using gene expression and fluorescent phospholipid based methodologies. Toxicol Sci. 2007 Sep;99(1):162-73.
|
| 21 |
Inverse agonist and neutral antagonist actions of antidepressants at recombinant and native 5-hydroxytryptamine2C receptors: differential modulatio... Mol Pharmacol. 2008 Mar;73(3):748-57.
|
| 22 |
Effects of selective serotonin reuptake inhibitors on three sex steroids in two versions of the aromatase enzyme inhibition assay and in the H295R cell assay. Toxicol In Vitro. 2015 Oct;29(7):1729-35.
|
| 23 |
Antidepressant drugs activate SREBP and up-regulate cholesterol and fatty acid biosynthesis in human glial cells. Neurosci Lett. 2006 Mar 13;395(3):185-90. doi: 10.1016/j.neulet.2005.10.096. Epub 2005 Dec 1.
|
| 24 |
Neuroendocrine effects of citalopram, a selective serotonin re-uptake inhibitor, during lifespan in humans. J Endocrinol Invest. 2010 Oct;33(9):657-62. doi: 10.1007/BF03346666. Epub 2010 Apr 22.
|
| 25 |
Cholinergic modulation of the cerebral metabolic response to citalopram in Alzheimer's disease. Brain. 2009 Feb;132(Pt 2):392-401. doi: 10.1093/brain/awn326. Epub 2009 Jan 19.
|
| 26 |
The antidepressants imipramine, clomipramine, and citalopram induce apoptosis in human acute myeloid leukemia HL-60 cells via caspase-3 activation. J Biochem Mol Toxicol. 1999;13(6):338-47. doi: 10.1002/(sici)1099-0461(1999)13:6<338::aid-jbt8>3.0.co;2-7.
|
| 27 |
Profiling of enantiopure drugs towards aryl hydrocarbon (AhR), glucocorticoid (GR) and pregnane X (PXR) receptors in human reporter cell lines. Chem Biol Interact. 2014 Feb 5;208:64-76. doi: 10.1016/j.cbi.2013.11.018. Epub 2013 Dec 6.
|
| 28 |
Antidepressant induced cholestasis: hepatocellular redistribution of multidrug resistant protein (MRP2). Gut. 2003 Feb;52(2):300-3. doi: 10.1136/gut.52.2.300.
|
| 29 |
Differential effects of psychoactive substances on human wildtype and polymorphic T356M dopamine transporters (DAT). Toxicology. 2019 Jun 15;422:69-75. doi: 10.1016/j.tox.2019.04.012. Epub 2019 Apr 19.
|
| 30 |
5-HTTLPR polymorphism of the serotonin transporter gene predicts non-remission in major depression patients treated with citalopram in a 12-weeks follow up study. J Clin Psychopharmacol. 2003 Dec;23(6):563-7. doi: 10.1097/01.jcp.0000095350.32154.73.
|
| 31 |
Association of GRIK4 with outcome of antidepressant treatment in the STAR*D cohort. Am J Psychiatry. 2007 Aug;164(8):1181-8. doi: 10.1176/appi.ajp.2007.06111790.
|
| 32 |
Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am J Hum Genet. 2006 May;78(5):804-814. doi: 10.1086/503820. Epub 2006 Mar 20.
|
| 33 |
Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 2008 Apr;78(1):91-102.
|
| 34 |
Nonclinical pharmacokinetics of oseltamivir and oseltamivir carboxylate in the central nervous system. Antimicrob Agents Chemother. 2009 Nov;53(11):4753-61.
|
| 35 |
Limited brain distribution of [3R,4R,5S]-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate phosphate (Ro 64-0802), a pharmacologically active form of oseltamivir, by active efflux across the blood-brain barrier mediated by organic anion transporter 3 (Oat3/Slc22a8) and multidrug resistance-associated protein 4 (Mrp4/Abcc4). Drug Metab Dispos. 2009 Feb;37(2):315-21.
|
| 36 |
Oseltamivir (tamiflu) is a substrate of peptide transporter 1. Drug Metab Dispos. 2009 Aug;37(8):1676-81.
|
| 37 |
FDA Drug Development and Drug Interactions
|
| 38 |
An in vitro coculture system of human peripheral blood mononuclear cells with hepatocellular carcinoma-derived cells for predicting drug-induced liver injury. Arch Toxicol. 2021 Jan;95(1):149-168. doi: 10.1007/s00204-020-02882-4. Epub 2020 Aug 20.
|
| 39 |
In vitro inhibition of carboxylesterase 1 by Kava (Piper methysticum) Kavalactones. Chem Biol Interact. 2022 Apr 25;357:109883. doi: 10.1016/j.cbi.2022.109883. Epub 2022 Mar 9.
|
| 40 |
Interleukin-6 alters the cellular responsiveness to clopidogrel, irinotecan, and oseltamivir by suppressing the expression of carboxylesterases HCE1 and HCE2. Mol Pharmacol. 2007 Sep;72(3):686-94. doi: 10.1124/mol.107.036889. Epub 2007 May 30.
|
|
|
|
|
|
|