| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
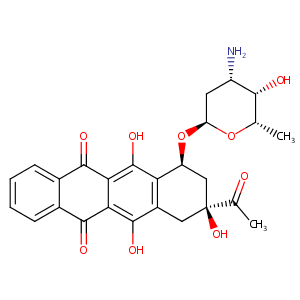
Idarubicin FDA Label
|
| 3 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7083).
|
| 4 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6793).
|
| 5 |
Quantitative high-throughput profiling of environmental chemicals and drugs that modulate farnesoid X receptor. Sci Rep. 2014 Sep 26;4:6437. doi: 10.1038/srep06437.
|
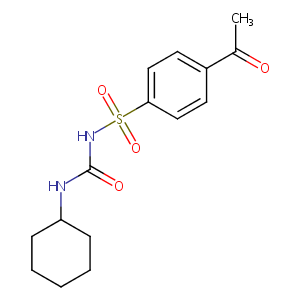
| 6 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services.
|
| 7 |
Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions. Pharm Res. 2013 Apr;30(4):996-1007.
|
| 8 |
Amonafide L-malate is not a substrate for multidrug resistance proteins in secondary acute myeloid leukemia. Leukemia. 2008 Nov;22(11):2110-5.
|
| 9 |
In vitro evaluation of cytochrome P450-mediated drug interactions between cytarabine, idarubicin, itraconazole and caspofungin. Hematology. 2004 Jun;9(3):217-21.
|
| 10 |
A Quantitative Approach to Screen for Nephrotoxic Compounds In Vitro. J Am Soc Nephrol. 2016 Apr;27(4):1015-28. doi: 10.1681/ASN.2015010060. Epub 2015 Aug 10.
|
| 11 |
The use of biochemical markers in cardiotoxicity monitoring in patients treated for leukemia. Neoplasma. 2005;52(5):430-4.
|
| 12 |
The induction of apoptosis by daunorubicin and idarubicin in human trisomic and diabetic fibroblasts. Cell Mol Biol Lett. 2008;13(2):182-94. doi: 10.2478/s11658-007-0045-7. Epub 2008 Apr 10.
|
| 13 |
Refining the human iPSC-cardiomyocyte arrhythmic risk assessment model. Toxicol Sci. 2013 Dec;136(2):581-94. doi: 10.1093/toxsci/kft205. Epub 2013 Sep 19.
|
| 14 |
Diabetes and insulin secretion: the ATP-sensitive K+ channel (K ATP) connection.Diabetes.2005 Nov;54(11):3065-72.
|
| 15 |
Clinically and pharmacologically relevant interactions of antidiabetic drugs. Ther Adv Endocrinol Metab. 2016 Apr;7(2):69-83.
|
| 16 |
Molecular and biochemical characterisation of human short-chain dehydrogenase/reductase member 3 (DHRS3). Chem Biol Interact. 2015 Jun 5;234:178-87.
|
| 17 |
Characterization of the binding of sulfonylurea drugs to HSA by high-performance affinity chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2010 Jun 1;878(19):1590-8. doi: 10.1016/j.jchromb.2010.04.019.
|
| 18 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
|
|
|
|
|
|