| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
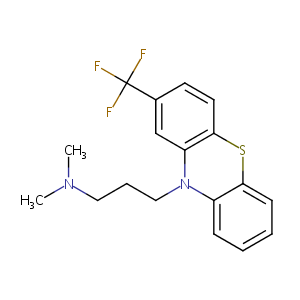
Triflupromazine FDA Label
|
| 3 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4330).
|
| 4 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015
|
| 5 |
Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection. Antimicrob Agents Chemother. 2014 Aug;58(8):4885-93.
|
| 6 |
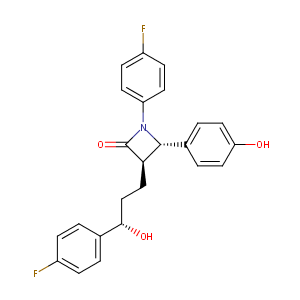
Ezetimibe FDA Label
|
| 7 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6816).
|
| 8 |
Some properties of 5-hydroxytryptamine receptors in the hindquarters of the rat. Br J Pharmacol. 1979 Sep;67(1):79-85.
|
| 9 |
Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions. Pharm Res. 2013 Apr;30(4):996-1007.
|
| 10 |
Why are most phospholipidosis inducers also hERG blockers?. Arch Toxicol. 2017 Dec;91(12):3885-3895. doi: 10.1007/s00204-017-1995-9. Epub 2017 May 27.
|
| 11 |
Identification of human UDP-glucuronosyltransferase enzyme(s) responsible for the glucuronidation of ezetimibe (Zetia). Drug Metab Dispos. 2004 Mar;32(3):314-20.
|
| 12 |
Update on patented cholesterol absorption inhibitors. Expert Opin Ther Pat. 2009 Aug;19(8):1083-107.
|
| 13 |
Intestinal expression of P-glycoprotein (ABCB1), multidrug resistance associated protein 2 (ABCC2), and uridine diphosphate-glucuronosyltransferase 1A1 predicts the disposition and modulates the effects of the cholesterol absorption inhibitor ezetimibe in humans. Clin Pharmacol Ther. 2006 Mar;79(3):206-17.
|
| 14 |
Pharmacokinetic and pharmacodynamic interactions between the immunosuppressant sirolimus and the lipid-lowering drug ezetimibe in healthy volunteers. Clin Pharmacol Ther. 2010 Jun;87(6):663-7.
|
| 15 |
Complex pharmacokinetic behavior of ezetimibe depends on abcc2, abcc3, and abcg2. Drug Metab Dispos. 2009 Aug;37(8):1698-702.
|
| 16 |
A LC-MS/MS method to quantify the novel cholesterol lowering drug ezetimibe in human serum, urine and feces in healthy subjects genotyped for SLCO1B1. J Chromatogr B Analyt Technol Biomed Life Sci. 2006 Jan 2;830(1):143-50.
|
| 17 |
Early identification of clinically relevant drug interactions with the human bile salt export pump (BSEP/ABCB11). Toxicol Sci. 2013 Dec;136(2):328-43.
|
| 18 |
Ezetimibe: a review of its metabolism, pharmacokinetics and drug interactions. Clin Pharmacokinet. 2005;44(5):467-94.
|
| 19 |
Drug interactions between the immunosuppressant tacrolimus and the cholesterol absorption inhibitor ezetimibe in healthy volunteers. Clin Pharmacol Ther. 2011 Apr;89(4):524-8.
|
| 20 |
Effects of ezetimibe and/or simvastatin on LDL receptor protein expression and on LDL receptor and HMG-CoA reductase gene expression: a randomized trial in healthy men. Atherosclerosis. 2008 May;198(1):198-207.
|
| 21 |
Carotenoid transport is decreased and expression of the lipid transporters SR-BI, NPC1L1, and ABCA1 is downregulated in Caco-2 cells treated with ezetimibe. J Nutr. 2005 Oct;135(10):2305-12. doi: 10.1093/jn/135.10.2305.
|
| 22 |
Compound heterozygosity for two non-synonymous polymorphisms in NPC1L1 in a non-responder to ezetimibe. Clin Genet. 2005 Feb;67(2):175-7. doi: 10.1111/j.1399-0004.2004.00388.x.
|
|
|
|
|
|
|