| Drug Name |
Decanal
|
| Synonyms |
Aldehyde C-10; Aldehyde C10; Capraldehyde; Capric aldehyde; Caprinaldehyde; Caprinic aldehyde; DECALDEHYDE; Decanal; Decanal (natural); Decanaldehyde; Decyl aldehyde; Decyl aldehyde, 95%; Decylic aldehyde; KSMVZQYAVGTKIV-UHFFFAOYSA-N; decylaldehyde; n-DECYLALDEHYDE; n-Decaldehyde; n-Decanal; n-Decyl aldehyde; 1-Decanal; 1-Decanal(mixed isomers); 1-Decyl aldehyde; 112-31-2; 31Z90Q7KQJ; AC1L1QFM; AI3-04860; BRN 1362530; C-10 aldehyde; CHEBI:31457; EINECS 203-957-4; FEMA No. 2362; HSDB 288; MFCD00007031; NSC 6087; NSC6087; UNII-31Z90Q7KQJ
|
| Structure |
|
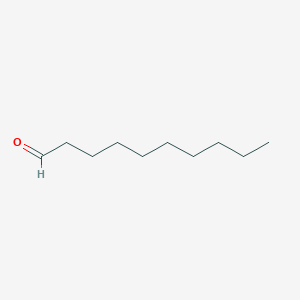 |
|
3D MOL
|
2D MOL
|
| #Ro5 Violations (Lipinski): 0 |
Molecular Weight (mw) |
156.26 |
|
| Logarithm of the Partition Coefficient (xlogp) |
3.8 |
| Rotatable Bond Count (rotbonds) |
8 |
| Hydrogen Bond Donor Count (hbonddonor) |
0 |
| Hydrogen Bond Acceptor Count (hbondacc) |
1 |
| Chemical Identifiers |
- Formula
- C10H20O
- IUPAC Name
decanal - Canonical SMILES
-
CCCCCCCCCC=O
- InChI
-
KSMVZQYAVGTKIV-UHFFFAOYSA-N
- InChIKey
-
1S/C10H20O/c1-2-3-4-5-6-7-8-9-10-11/h10H,2-9H2,1H3
|
| Cross-matching ID |
- PubChem CID
- 8175
- ChEBI ID
-
- CAS Number
-
- INTEDE ID
- DR2033
|
|
|
|
|
|
|
|



