| Drug Name |
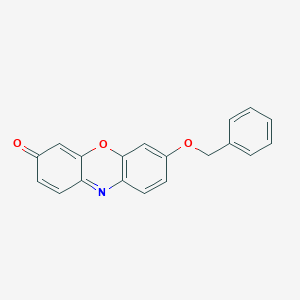
Benzyloxyresorufin
|
| Synonyms |
Benzyloxyresorfin; 7-Benzyloxyresorufin; J-100240; MCULE-6922850124; O7-Benzylresorufin; Resorufin benzyl ether; SCHEMBL563335; STK299141; ZINC4534147; 3H-Phenoxazin-3-one, 7-(phenylmethoxy)-; 7-(Phenylmethoxy)-3H-phenoxazin-3-one; 7-(benzyloxy)-3H-phenoxazin-3-one; 7-Benzoxyresorufin; 7-Benzyloxy-3H-phenoxazin-3-one; 87687-02-3; AKOS005429005; CTK8D9518; DTXSID90236553; FT-0642066; MFCD00171613; Resorufin benzyl ether CYP450 substrate; Resorufin benzyl ether, CYP450 substrate
|
| Structure |
|
 |
|
3D MOL
|
2D MOL
|
| #Ro5 Violations (Lipinski): 0 |
Molecular Weight (mw) |
303.3 |
|
| Logarithm of the Partition Coefficient (xlogp) |
2.9 |
| Rotatable Bond Count (rotbonds) |
3 |
| Hydrogen Bond Donor Count (hbonddonor) |
0 |
| Hydrogen Bond Acceptor Count (hbondacc) |
4 |
| Chemical Identifiers |
- Formula
- C19H13NO3
- IUPAC Name
7-phenylmethoxyphenoxazin-3-one - Canonical SMILES
-
C1=CC=C(C=C1)COC2=CC3=C(C=C2)N=C4C=CC(=O)C=C4O3
- InChI
-
XNZRYTITWLGTJS-UHFFFAOYSA-N
- InChIKey
-
1S/C19H13NO3/c21-14-6-8-16-18(10-14)23-19-11-15(7-9-17(19)20-16)22-12-13-4-2-1-3-5-13/h1-11H,12H2
|
| Cross-matching ID |
- PubChem CID
- 114982
- CAS Number
-
- INTEDE ID
- DR2451
|
|
|
|
|
|
|
|



