Details of the Drug
General Information of Drug (ID: DMUNK59)
| Drug Name |
Bafilomycin A1
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
bafilomycin A1; 88899-55-2; MFCD06795130; CHEMBL290814; CHEBI:22689; (3Z,5E,7R,8S,9S,11E,13E,15S,16R)-16-{(2S,3R,4S)-4-[(2R,4R,5S,6R)-2,4-dihydroxy-6-isopropyl-5-methyltetrahydro-2H-pyran-2-yl]-3-hydroxypentan-2-yl}-8-hydroxy-3,15-dimethoxy-5,7,9,11-tetramethyloxacyclohexadeca-3,5,11,13-tetraen-2-one; Bafilomycin; NSC381866; Bafilomycin A1/; Hygrolidin, 21-O-de(3-carboxy-1-oxo-2-propenyl)-2-demethyl-2-methoxy-24-methyl-; Bafilomycin A1 from Streptomyces griseus; BSPBio_001470; MEGxm0_000385; SCHEMBL13775181; ACon0_000813; HMS3402J12; Bafilomycin A1 Ready Made Solution; ABP000610; BDBM50064186; AKOS030213158; ZINC169647947; DB06733; MCULE-2359469972; NCGC00163426-02; (3Z,5E,7R,8S,9S,11E,13E,15S,16R)-16-[(2S,3R,4S)-4-[(2R,4R,5S,6R)-2,4-dihydroxy-5-methyl-6-propan-2-yloxan-2-yl]-3-hydroxypentan-2-yl]-8-hydroxy-3,15-dimethoxy-5,7,9,11-tetramethyl-1-oxacyclohexadeca-3,5,11,13-tetraen-2-one; Q4841341; (3Z,5E,7R,8S,9S,11E,13E,15S,16R)-16-[(1S,2R,3S)-3-[(2R,4R,5S,6R)-2,4-dihydroxy-6-isopropyl-5-methyl-tetrahydropyran-2-yl]-2-hydroxy-1-methyl-butyl]-8-hydroxy-3,15-dimethoxy-5,7,9,11-tetramethyl-1-oxacyclohexadeca-3,5,11,13-tetraen-2-one
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
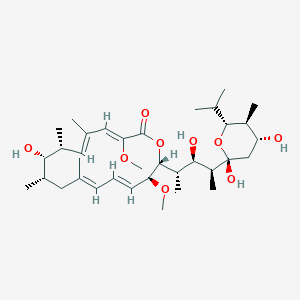 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 2 | Molecular Weight (mw) | 622.8 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 6 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 7 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 4 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 9 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
| Combinatorial Drugs (CBD) | Click to Jump to the Detailed CBD Information of This Drug | ||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Experimental Cancer Drug Sensitivity Information
References
| 1 | Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica. 2015 Mar;100(3):345-56. | ||||
|---|---|---|---|---|---|
| 2 | Molecular basis of V-ATPase inhibition by bafilomycin A1. Nat Commun. 2021 Mar 19;12(1):1782. | ||||
| 3 | Chronic high glucose inhibits albumin reabsorption by lysosomal alkalinization in cultured porcine proximal tubular epithelial cells (LLC-PK1). Diabetes Res Clin Pract. 2006 Jun;72(3):223-30. doi: 10.1016/j.diabres.2005.10.019. Epub 2005 Nov 28. | ||||
| 4 | Nano-sized iron particles may induce multiple pathways of cell death following generation of mistranscripted RNA in human corneal epithelial cells. Toxicol In Vitro. 2017 Aug;42:348-357. | ||||
| 5 | Combined effects of EGFR tyrosine kinase inhibitors and vATPase inhibitors in NSCLC cells. Toxicol Appl Pharmacol. 2015 Aug 15;287(1):17-25. doi: 10.1016/j.taap.2015.05.001. Epub 2015 May 14. | ||||
| 6 | Inhibition of cholesterol metabolism underlies synergy between mTOR pathway inhibition and chloroquine in bladder cancer cells. Oncogene. 2016 Aug 25;35(34):4518-28. doi: 10.1038/onc.2015.511. Epub 2016 Feb 8. | ||||
| 7 | Autophagy induction by capsaicin in malignant human breast cells is modulated by p38 and extracellular signal-regulated mitogen-activated protein kinases and retards cell death by suppressing endoplasmic reticulum stress-mediated apoptosis. Mol Pharmacol. 2010 Jul;78(1):114-25. doi: 10.1124/mol.110.063495. Epub 2010 Apr 6. | ||||
| 8 | Berbamine Hydrochloride inhibits lysosomal acidification by activating Nox2 to potentiate chemotherapy-induced apoptosis via the ROS-MAPK pathway in human lung carcinoma cells. Cell Biol Toxicol. 2023 Aug;39(4):1297-1317. doi: 10.1007/s10565-022-09756-8. Epub 2022 Sep 7. | ||||
| 9 | GJA1 reverses arsenic-induced EMT via modulating MAPK/ERK signaling pathway. Toxicol Appl Pharmacol. 2022 Sep 1;450:116138. doi: 10.1016/j.taap.2022.116138. Epub 2022 Jun 21. | ||||
| 10 | SB202190 inhibits endothelial cell apoptosis via induction of autophagy and heme oxygenase-1. Oncotarget. 2018 May 1;9(33):23149-23163. doi: 10.18632/oncotarget.25234. eCollection 2018 May 1. | ||||
