Details of the Drug
General Information of Drug (ID: DMNI3U2)
| Drug Name |
ZIMELIDINE
|
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
Prestwick0_000092; Prestwick1_000092; 56775-88-3; AC1L26VA; NCIStruc1_001846; NCIStruc2_001359; SPBio_001983; 2-Propen-1-amine, 3-(4-bromophenyl)-N,N-dimethyl-3-(3-pyridinyl)-; CTK1F3832; OYPPVKRFBIWMSX-UHFFFAOYSA-N; DB04832; NCI60_002580; FT-0675907; N,N-dimethyl-3-(4-bromophenyl)-3-(pyrid-3-yl)allylamine; N,N-dimethyl-3-(4-bromophenyl)-3-(3-pyridyl)-allylamine; (Z)-3-[1-(p-Bromophenyl)-3-(dimethylamino)propenyl]pyridine; 3-(4-bromophenyl)-N,N-dimethyl-3-pyridin-3-ylprop-2-en-1-amine
|
||||||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||||||
| Affected Organisms |
Humans and other mammals
|
||||||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||||||
| Structure |
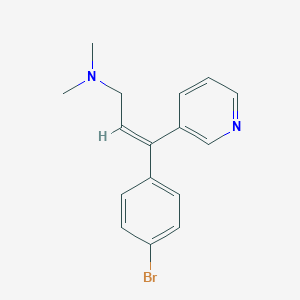 |
||||||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 317.22 | |||||||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 3.9 | ||||||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 4 | ||||||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 0 | ||||||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 2 | ||||||||||||||||||||||||||
| ADMET Property |
|
||||||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| ICD Disease Classification | 06 Mental, behavioural or neurodevelopmental disorder | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease Class | ICD-11: 6A20 Schizophrenia | |||||||||||||||||||||||
| The Studied Tissue | Pre-frontal cortex | |||||||||||||||||||||||
| The Studied Disease | Major depressive disorder [ICD-11:6A20] | |||||||||||||||||||||||
|
||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||
References
