Details of the Drug
General Information of Drug (ID: DMW16X2)
| Drug Name |
UDP-glucuronic acid
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms | UDP glucuronic acid; UDP-glucuronate; UDPGA; uridine diphosphate glucuronic acid | ||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
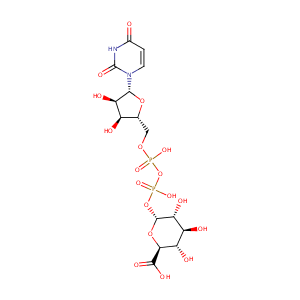 |
||||||||||||||||||||||
| 3D MOL is unavailable | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 4 | Molecular Weight (mw) | 580.28 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | -6.4 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 9 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 9 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 18 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug-Metabolizing Enzyme (DME) |
|
||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | |||||||||||||||||||||||||||||||||||||
References
