| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
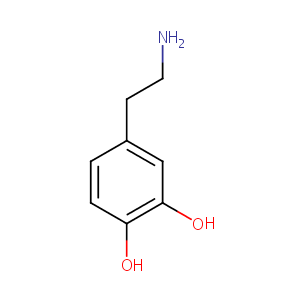
Dopamine FDA Label
|
| 3 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 940).
|
| 4 |
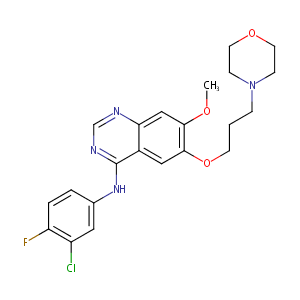
Gefitinib FDA Label
|
| 5 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4941).
|
| 6 |
Mitochondrial proteomics investigation of a cellular model of impaired dopamine homeostasis, an early step in Parkinson's disease pathogenesis. Mol Biosyst. 2014 Jun;10(6):1332-44.
|
| 7 |
Vitamin D signaling and the differentiation of developing dopamine systems. Neuroscience. 2016 Oct 1;333:193-203. doi: 10.1016/j.neuroscience.2016.07.020. Epub 2016 Jul 20.
|
| 8 |
Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol Psychiatry. 2011 Sep;16(9):927-37, 881. doi: 10.1038/mp.2011.32. Epub 2011 Apr 19.
|
| 9 |
The Detection of Dopamine Gene Receptors (DRD1-DRD5) Expression on Human Peripheral Blood Lymphocytes by Real Time PCR. Iran J Allergy Asthma Immunol. 2004 Dec;3(4):169-74.
|
| 10 |
Differential pharmacological in vitro properties of organic cation transporters and regional distribution in rat brain. Neuropharmacology. 2006 Jun;50(8):941-52.
|
| 11 |
Organic cation transporters and their pharmacokinetic and pharmacodynamic consequences. Drug Metab Pharmacokinet. 2008;23(4):243-53.
|
| 12 |
SLC18: Vesicular neurotransmitter transporters for monoamines and acetylcholine. Mol Aspects Med. 2013 Apr-Jun;34(2-3):360-72.
|
| 13 |
Synaptic vesicle glycoprotein 2C (SV2C) modulates dopamine release and is disrupted in Parkinson disease. Proc Natl Acad Sci U S A. 2017 Mar 14;114(11):E2253-E2262.
|
| 14 |
Characterization of VNTRs Within the Entire Region of SLC6A3 and Its Association with Hypertension. DNA Cell Biol. 2017 Mar;36(3):227-236.
|
| 15 |
Modulation of CYP1A2 enzyme activity by indoleamines: inhibition by serotonin and tryptamine. Pharmacogenetics. 1998 Jun;8(3):251-8.
|
| 16 |
Pharmacogenetics of schizophrenia. Am J Med Genet. 2000 Spring;97(1):98-106.
|
| 17 |
Association between polymorphisms in catechol-O-methyltransferase (COMT) and cocaine-induced paranoia in European-American and African-American populations. Am J Med Genet B Neuropsychiatr Genet. 2011 Sep;156B(6):651-60.
|
| 18 |
Monoamine oxidases (MAO) in the pathogenesis of heart failure and ischemia/reperfusion injury. Biochim Biophys Acta. 2011 Jul;1813(7):1323-32.
|
| 19 |
Molecular cloning, expression, and functional characterization of novel mouse sulfotransferases. Biochem Biophys Res Commun. 1998 Jun 29;247(3):681-6.
|
| 20 |
Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019 Jun 14;364(6445). pii: eaau6323.
|
| 21 |
Effect of penicillin-based antibiotics, amoxicillin, ampicillin, and piperacillin, on drug-metabolizing activities of human hepatic cytochromes P450. J Toxicol Sci. 2016 Feb;41(1):143-6.
|
| 22 |
Inhibition potential of 3,4-methylenedioxymethamphetamine (MDMA) and its metabolites on the in vitro monoamine oxidase (MAO)-catalyzed deamination of the neurotransmitters serotonin and dopamine. Toxicol Lett. 2016 Jan 22;243:48-55.
|
| 23 |
Molecular mechanisms controlling the rate and specificity of catechol O-methylation by human soluble catechol O-methyltransferase. Mol Pharmacol. 2001 Feb;59(2):393-402. doi: 10.1124/mol.59.2.393.
|
| 24 |
Functional characterization of N-octyl 4-methylamphetamine variants and related bivalent compounds at the dopamine and serotonin transporters using Ca(2+) channels as sensors. Toxicol Appl Pharmacol. 2021 May 15;419:115513. doi: 10.1016/j.taap.2021.115513. Epub 2021 Mar 27.
|
| 25 |
The effect of rare human sequence variants on the function of vesicular monoamine transporter 2. Pharmacogenetics. 2004 Sep;14(9):587-94. doi: 10.1097/00008571-200409000-00003.
|
| 26 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
| 27 |
Effects of dopamine on LC3-II activation as a marker of autophagy in a neuroblastoma cell model. Neurotoxicology. 2009 Jul;30(4):658-65. doi: 10.1016/j.neuro.2009.04.007. Epub 2009 May 4.
|
| 28 |
Inhibition of human glutathione S-transferases by dopamine, alpha-methyldopa and their 5-S-glutathionyl conjugates. Chem Biol Interact. 1994 Jan;90(1):87-99.
|
| 29 |
Activation of methionine synthase by insulin-like growth factor-1 and dopamine: a target for neurodevelopmental toxins and thimerosal. Mol Psychiatry. 2004 Apr;9(4):358-70.
|
| 30 |
Functional expression and comparative characterization of nine murine cytochromes P450 by fluorescent inhibition screening. Drug Metab Dispos. 2008 Jul;36(7):1322-31.
|
| 31 |
Induction of parkin expression in the presence of oxidative stress. Eur J Neurosci. 2006 Sep;24(5):1366-72. doi: 10.1111/j.1460-9568.2006.04998.x.
|
| 32 |
Mitochondrial UCP5 is neuroprotective by preserving mitochondrial membrane potential, ATP levels, and reducing oxidative stress in MPP+ and dopamine toxicity. Free Radic Biol Med. 2010 Sep 15;49(6):1023-35. doi: 10.1016/j.freeradbiomed.2010.06.017. Epub 2010 Jun 19.
|
| 33 |
Ligand binding and aggregation of pathogenic SOD1. Nat Commun. 2013;4:1758. doi: 10.1038/ncomms2750.
|
| 34 |
Dose-dependent separation of dopaminergic and adrenergic effects of epinine in healthy volunteers. Naunyn Schmiedebergs Arch Pharmacol. 1995 Oct;352(4):429-37. doi: 10.1007/BF00172781.
|
| 35 |
Effect of drugs interacting with the dopaminergic receptors on glucose levels and insulin release in healthy and type 2 diabetic subjects. Am J Ther. 2008 Jul-Aug;15(4):397-402. doi: 10.1097/MJT.0b013e318160c353.
|
| 36 |
Parkin protects human dopaminergic neuroblastoma cells against dopamine-induced apoptosis. Hum Mol Genet. 2004 Aug 15;13(16):1745-54. doi: 10.1093/hmg/ddh180. Epub 2004 Jun 15.
|
| 37 |
Resveratrol protects SH-SY5Y neuroblastoma cells from apoptosis induced by dopamine. Exp Mol Med. 2007 Jun 30;39(3):376-84. doi: 10.1038/emm.2007.42.
|
| 38 |
Characterizing fucoxanthin as a selective dopamine D(3)/D(4) receptor agonist: Relevance to Parkinson's disease. Chem Biol Interact. 2019 Sep 1;310:108757. doi: 10.1016/j.cbi.2019.108757. Epub 2019 Jul 16.
|
| 39 |
Dopamine down-regulation of protein L-isoaspartyl methyltransferase is dependent on reactive oxygen species in SH-SY5Y cells. Neuroscience. 2014 May 16;267:263-76. doi: 10.1016/j.neuroscience.2014.03.001. Epub 2014 Mar 12.
|
| 40 |
Sulfation of environmental estrogen-like chemicals by human cytosolic sulfotransferases. Biochem Biophys Res Commun. 2000 Jan 7;267(1):80-4. doi: 10.1006/bbrc.1999.1935.
|
| 41 |
Role of tumor necrosis factor-alpha in methamphetamine-induced drug dependence and neurotoxicity. J Neurosci. 2004 Mar 3;24(9):2212-25. doi: 10.1523/JNEUROSCI.4847-03.2004.
|
| 42 |
Expression of tyrosine hydroxylase increases the resistance of human neuroblastoma cells to oxidative insults. Toxicol Sci. 2010 Jan;113(1):150-7. doi: 10.1093/toxsci/kfp245. Epub 2009 Oct 8.
|
| 43 |
G209A mutant alpha synuclein expression specifically enhances dopamine induced oxidative damage. Neurochem Int. 2004 Oct;45(5):669-76. doi: 10.1016/j.neuint.2004.03.029.
|
| 44 |
Increased arterial pressure in mice with overexpression of the ADHD candidate gene calcyon in forebrain. PLoS One. 2019 Feb 12;14(2):e0211903. doi: 10.1371/journal.pone.0211903. eCollection 2019.
|
| 45 |
Selective transport of monoamine neurotransmitters by human plasma membrane monoamine transporter and organic cation transporter 3. J Pharmacol Exp Ther. 2010 Dec;335(3):743-53. doi: 10.1124/jpet.110.170142. Epub 2010 Sep 21.
|
| 46 |
Administration of secretin for autism alters dopamine metabolism in the central nervous system. Brain Dev. 2006 Mar;28(2):99-103. doi: 10.1016/j.braindev.2005.05.005. Epub 2005 Sep 15.
|
| 47 |
Synergistic antitumor effect of S-1 and the epidermal growth factor receptor inhibitor gefitinib in non-small cell lung cancer cell lines: role of gefitinib-induced down-regulation of thymidylate synthase. Mol Cancer Ther. 2008 Mar;7(3):599-606.
|
| 48 |
Frankincense myrrh attenuates hepatocellular carcinoma by regulating tumor blood vessel development through multiple epidermal growth factor receptor-mediated signaling pathways. World J Gastrointest Oncol. 2022 Feb 15;14(2):450-477. doi: 10.4251/wjgo.v14.i2.450.
|
| 49 |
EGFR inhibitors enhanced the susceptibility to NK cell-mediated lysis of lung cancer cells. J Immunother. 2011 May;34(4):372-81. doi: 10.1097/CJI.0b013e31821b724a.
|
| 50 |
Overcoming acquired resistance of gefitinib in lung cancer cells without T790M by AZD9291 or Twist1 knockdown in vitro and in vivo. Arch Toxicol. 2019 Jun;93(6):1555-1571. doi: 10.1007/s00204-019-02453-2. Epub 2019 Apr 16.
|
| 51 |
DNA repair gene polymorphisms and benefit from gefitinib in never-smokers with lung adenocarcinoma. Cancer. 2011 Jul 15;117(14):3201-8. doi: 10.1002/cncr.25863. Epub 2011 Jan 24.
|
| 52 |
Gefitinib ('Iressa', ZD1839) and new epidermal growth factor receptor inhibitors. Br J Cancer. 2004 Feb 9;90(3):566-72.
|
| 53 |
Gefitinib-phenytoin interaction is not correlated with the C-erythromycin breath test in healthy male volunteers. Br J Clin Pharmacol. 2009 Aug;68(2):226-37.
|
| 54 |
Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016 Jan 1;370(1):153-64.
|
| 55 |
Contribution of OATP1B1 and OATP1B3 to the disposition of sorafenib and sorafenib-glucuronide. Clin Cancer Res. 2013 Mar 15;19(6):1458-66.
|
| 56 |
Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin Cancer Res. 2007 Jun 15;13(12):3731-7.
|
| 57 |
Pharmacokinetic drug interactions of gefitinib with rifampicin, itraconazole and metoprolol. Clin Pharmacokinet. 2005;44(10):1067-81.
|
| 58 |
Pharmacogenetics of ABCG2 and adverse reactions to gefitinib. J Natl Cancer Inst. 2006 Dec 6;98(23):1739-42.
|
| 59 |
Dasatinib (BMS-354825) selectively induces apoptosis in lung cancer cells dependent on epidermal growth factor receptor signaling for survival. Cancer Res. 2006 Jun 1;66(11):5542-8. doi: 10.1158/0008-5472.CAN-05-4620.
|
| 60 |
Identification of genes linked to gefitinib treatment in prostate cancer cell lines with or without resistance to androgen: a clue to application of gefitinib to hormone-resistant prostate cancer. Oncol Rep. 2006 Jun;15(6):1453-60.
|
| 61 |
Interference with bile salt export pump function is a susceptibility factor for human liver injury in drug development. Toxicol Sci. 2010 Dec; 118(2):485-500.
|
| 62 |
Effects and mechanisms of betulinic acid on improving EGFR TKI-resistance of lung cancer cells. Environ Toxicol. 2018 Nov;33(11):1153-1159.
|
| 63 |
Antiproliferative effects of gefitinib are associated with suppression of E2F-1 expression and telomerase activity. Anticancer Res. 2006 Sep-Oct;26(5A):3387-91.
|
| 64 |
Leptomycin B reduces primary and acquired resistance of gefitinib in lung cancer cells. Toxicol Appl Pharmacol. 2017 Nov 15;335:16-27. doi: 10.1016/j.taap.2017.09.017. Epub 2017 Sep 21.
|
| 65 |
EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6(6):e18691. doi: 10.1371/journal.pone.0018691. Epub 2011 Jun 2.
|
| 66 |
ZD1839 induces p15INK4b and causes G1 arrest by inhibiting the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway. Mol Cancer Ther. 2007 May;6(5):1579-87. doi: 10.1158/1535-7163.MCT-06-0814.
|
| 67 |
Reactive metabolite of gefitinib activates inflammasomes: implications for gefitinib-induced idiosyncratic reaction. J Toxicol Sci. 2020;45(11):673-680. doi: 10.2131/jts.45.673.
|
| 68 |
Systems pharmacological analysis of drugs inducing stevens-johnson syndrome and toxic epidermal necrolysis. Chem Res Toxicol. 2015 May 18;28(5):927-34. doi: 10.1021/tx5005248. Epub 2015 Apr 3.
|
| 69 |
Bidirectional cross talk between ERalpha and EGFR signalling pathways regulates tamoxifen-resistant growth. Breast Cancer Res Treat. 2006 Mar;96(2):131-46. doi: 10.1007/s10549-005-9070-2. Epub 2005 Oct 27.
|
| 70 |
Epidermal growth factor receptor activity determines response of colorectal cancer cells to gefitinib alone and in combination with chemotherapy. Clin Cancer Res. 2005 Oct 15;11(20):7480-9. doi: 10.1158/1078-0432.CCR-05-0328.
|
| 71 |
Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007 May 1;13(9):2795-803. doi: 10.1158/1078-0432.CCR-06-2077.
|
| 72 |
Evidence of securin-mediated resistance to gefitinib-induced apoptosis in human cancer cells. Chem Biol Interact. 2013 Apr 25;203(2):412-22. doi: 10.1016/j.cbi.2013.03.011. Epub 2013 Mar 22.
|
| 73 |
Crosstalk between alveolar macrophages and alveolar epithelial cells/fibroblasts contributes to the pulmonary toxicity of gefitinib. Toxicol Lett. 2021 Mar 1;338:1-9. doi: 10.1016/j.toxlet.2020.11.011. Epub 2020 Nov 25.
|
| 74 |
Cytotoxicity of 34 FDA approved small-molecule kinase inhibitors in primary rat and human hepatocytes. Toxicol Lett. 2018 Jul;291:138-148. doi: 10.1016/j.toxlet.2018.04.010. Epub 2018 Apr 12.
|
| 75 |
Growth of hormone-dependent MCF-7 breast cancer cells is promoted by constitutive caveolin-1 whose expression is lost in an EGF-R-mediated manner during development of tamoxifen resistance. Breast Cancer Res Treat. 2010 Feb;119(3):575-91. doi: 10.1007/s10549-009-0355-8. Epub 2009 Mar 15.
|
| 76 |
Dihydromyricetin suppresses tumor growth via downregulation of the EGFR/Akt/survivin signaling pathway. J Biochem Mol Toxicol. 2023 Jun;37(6):e23328. doi: 10.1002/jbt.23328. Epub 2023 Feb 19.
|
| 77 |
The anti-cancer drug gefitinib accelerates Fas-mediated apoptosis by enhancing caspase-8 activation in cancer cells. J Toxicol Sci. 2019;44(6):435-440. doi: 10.2131/jts.44.435.
|
| 78 |
Autophagy Inhibition Overcomes the Antagonistic Effect Between Gefitinib and Cisplatin in Epidermal Growth Factor Receptor Mutant Non--Small-Cell Lung Cancer Cells. Clin Lung Cancer. 2015 Sep;16(5):e55-66. doi: 10.1016/j.cllc.2015.03.006. Epub 2015 Apr 2.
|
| 79 |
Nrf2 but not autophagy inhibition is associated with the survival of wild-type epidermal growth factor receptor non-small cell lung cancer cells. Toxicol Appl Pharmacol. 2016 Nov 1;310:140-149. doi: 10.1016/j.taap.2016.09.010. Epub 2016 Sep 14.
|
| 80 |
Association of CYP1A1 and CYP1B1 inhibition in in vitro assays with drug-induced liver injury. J Toxicol Sci. 2021;46(4):167-176. doi: 10.2131/jts.46.167.
|
| 81 |
Susceptibility to natural killer cell-mediated lysis of colon cancer cells is enhanced by treatment with epidermal growth factor receptor inhibitors through UL16-binding protein-1 induction. Cancer Sci. 2012 Jan;103(1):7-16. doi: 10.1111/j.1349-7006.2011.02109.x. Epub 2011 Nov 15.
|
| 82 |
Combined tamoxifen and gefitinib in non-small cell lung cancer shows antiproliferative effects. Biomed Pharmacother. 2010 Feb;64(2):88-92. doi: 10.1016/j.biopha.2009.06.010. Epub 2009 Oct 23.
|
| 83 |
Inorganic arsenic exposure promotes malignant progression by HDAC6-mediated down-regulation of HTRA1. J Appl Toxicol. 2023 Aug;43(8):1214-1224. doi: 10.1002/jat.4457. Epub 2023 Mar 11.
|
| 84 |
A high-throughput screen for teratogens using human pluripotent stem cells. Toxicol Sci. 2014 Jan;137(1):76-90. doi: 10.1093/toxsci/kft239. Epub 2013 Oct 23.
|
| 85 |
Inhibition of SREBP increases gefitinib sensitivity in non-small cell lung cancer cells. Oncotarget. 2016 Aug 9;7(32):52392-52403.
|
| 86 |
The K-Ras effector p38 MAPK confers intrinsic resistance to tyrosine kinase inhibitors by stimulating EGFR transcription and EGFR dephosphorylation. J Biol Chem. 2017 Sep 8;292(36):15070-15079. doi: 10.1074/jbc.M117.779488. Epub 2017 Jul 24.
|
| 87 |
Slug confers resistance to the epidermal growth factor receptor tyrosine kinase inhibitor. Am J Respir Crit Care Med. 2011 Apr 15;183(8):1071-9. doi: 10.1164/rccm.201009-1440OC. Epub 2010 Oct 29.
|
| 88 |
Identification of protein expression alterations in gefitinib-resistant human lung adenocarcinoma: PCNT and mPR play key roles in the development of gefitinib-associated resistance. Toxicol Appl Pharmacol. 2015 Nov 1;288(3):359-73. doi: 10.1016/j.taap.2015.08.008. Epub 2015 Aug 20.
|
| 89 |
Induction of CYP1A1 increases gefitinib-induced oxidative stress and apoptosis in A549 cells. Toxicol In Vitro. 2017 Oct;44:36-43.
|
| 90 |
Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839). Hum Mol Genet. 2004 Dec 15;13(24):3029-43. doi: 10.1093/hmg/ddh331. Epub 2004 Oct 20.
|
| 91 |
The transcription factor FOXO3a is a crucial cellular target of gefitinib (Iressa) in breast cancer cells. Mol Cancer Ther. 2007 Dec;6(12 Pt 1):3169-79. doi: 10.1158/1535-7163.MCT-07-0507.
|
| 92 |
Aurora-A promotes gefitinib resistance via a NF-B signaling pathway in p53 knockdown lung cancer cells. Biochem Biophys Res Commun. 2011 Feb 11;405(2):168-72. doi: 10.1016/j.bbrc.2011.01.001. Epub 2011 Jan 7.
|
| 93 |
BIM induction of apoptosis triggered by EGFR-sensitive and resistance cell lines of non-small-cell lung cancer. Med Oncol. 2011 Jun;28(2):572-7. doi: 10.1007/s12032-010-9470-y. Epub 2010 Mar 17.
|
| 94 |
E3 ubiquitin ligase RNF180 reduces sensitivity of triple-negative breast cancer cells to Gefitinib by downregulating RAD51. Chem Biol Interact. 2022 Feb 25;354:109798. doi: 10.1016/j.cbi.2022.109798. Epub 2022 Jan 6.
|
| 95 |
Increases of amphiregulin and transforming growth factor-alpha in serum as predictors of poor response to gefitinib among patients with advanced non-small cell lung cancers. Cancer Res. 2005 Oct 15;65(20):9176-84. doi: 10.1158/0008-5472.CAN-05-1556.
|
|
|
|
|
|
|