| Synonyms |
beta-maltose; Maltobiose; Maltose, pure; Advanctose 100; 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranose; Cextromaltose; Maltodiose; Finetose; Sunmalt; Malt sugar; Finetose F; Sunmalt S; UNII-R4B6462NGR; D-Glucose, 4-O-alpha-D-glucopyranosyl-; AI3-09089; EINECS 200-716-5; BRN 0093798; 4-O-alpha-D-Glucopyranosyl-D-glucose; R4B6462NGR; 4-(alpha-D-Glucosido)-D-glucose; CHEBI:18147; beta-D-Cellobiose; GUBGYTABKSRVRQ-QUYVBRFLSA-N; beta-D-glucopyranose, 4-O-alpha-D-glucopyranosyl-
|
| Chemical Identifiers |
- Formula
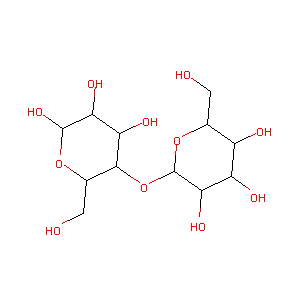
- C12H22O11
- IUPAC Name
(2R,3S,4S,5R,6R)-2-(hydroxymethyl)-6-[(2R,3S,4R,5R,6R)-4,5,6-trihydroxy-2-(hydroxymethyl)oxan-3-yl]oxyoxane-3,4,5-triol - Canonical SMILES
-
C([C@@H]1[C@H]([C@@H]([C@H]([C@H](O1)O[C@@H]2[C@H](O[C@H]([C@@H]([C@H]2O)O)O)CO)O)O)O)O
- InChI
-
InChI=1S/C12H22O11/c13-1-3-5(15)6(16)9(19)12(22-3)23-10-4(2-14)21-11(20)8(18)7(10)17/h3-20H,1-2H2/t3-,4-,5-,6+,7-,8-,9-,10-,11-,12-/m1/s1
- InChIKey
-
GUBGYTABKSRVRQ-QUYVBRFLSA-N
|