Details of the Drug
General Information of Drug (ID: DMINO7Z)
| Drug Name |
Perhexiline
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
Perhexilene; Perhexilina; Perhexilinum; Perhexilline; Perhexilina [INN-Spanish]; Perhexiline (INN); Perhexiline [INN:BAN]; Perhexilinum [INN-Latin]; (+)-2-(2,2-Dicyclohexylethyl)piperidine; (-)-2-(2,2-Dicyclohexylethyl)piperidine; 2-(2,2-Dicyclohexylethyl)piperidine
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Therapeutic Class |
Vasodilator Agents
|
||||||||||||||||||||||
| Affected Organisms |
Humans and other mammals
|
||||||||||||||||||||||
| ATC Code | |||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
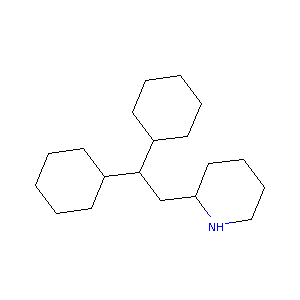 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 1 | Molecular Weight (mw) | 277.5 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 6.8 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 4 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 1 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 1 | ||||||||||||||||||||||
| ADMET Property |
|
||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
| Combinatorial Drugs (CBD) | Click to Jump to the Detailed CBD Information of This Drug | ||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
