Details of the Drug
General Information of Drug (ID: DMSJDTY)
| Drug Name |
Menadione
|
|||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
menadione; 58-27-5; Vitamin K3; 2-Methyl-1,4-naphthoquinone; Menaphthone; 2-methylnaphthalene-1,4-dione; 2-Methylnaphthoquinone; Thyloquinone; Panosine; Klottone; Kayquinone; Kappaxin; Kayklot; Kolklot; 2-Methyl-1,4-naphthalenedione; Menaphthon; Menadion; Kipca; Kanone; Mitenone; Kativ-G; Kaergona; Aquinone; Prokayvit; Mitenon; Koaxin; Kaykot; Kareon; Hemodal; Aquakay; Synkay; Karcon; K-Thrombyl; K-Vitan; Kipca-Oil Soluble; Juva-K; Vitamin K2(0); Vitamin K0; Menaquinone 0; Vitamin K 3; Menaphtone; Menadionum; 2-Methyl-1,4-naphthochinon; Usaf; Aquakay; Klottone;Koaxin; Memodol; Methylnaphthoquinone; Vicasol; Menadione semiquinone; M0373; VK3; Kappaxan (VAN); Kappaxin (TN); Kipca, oil soluble; Menadione (K3); Menadione (USP); Menadione [USAN:BAN]; Usaf ek-5185; Vitamin-K3; Methyl-1,4-naphthalenedione; Methyl-1,4-naphthoquinone; Vitamin K3 : 2-Methyl-1,4-naphthoquinone; 1,4-Naphthalenedione, 2-methyl-,radical ion(1-); 2-Methyl-1,4-naftochinon; 2-Methyl-1,4-naftochinon [Czech]; 2-Methyl-1,4-naphthochinon [German]; 2-methyl-1,4-naphthoquinone, 5; 3-Methyl-1,4-naphthoquinone; Ascorbic acid/menadione; Triglycyl-lysine-vasopressin
|
|||||||||||||||||||||||||||||||||||||||||||||
| Indication |
|
|||||||||||||||||||||||||||||||||||||||||||||
| Therapeutic Class |
Vitamins
|
|||||||||||||||||||||||||||||||||||||||||||||
| Affected Organisms |
Humans and other mammals
|
|||||||||||||||||||||||||||||||||||||||||||||
| ATC Code | ||||||||||||||||||||||||||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
|||||||||||||||||||||||||||||||||||||||||||||
| Structure |
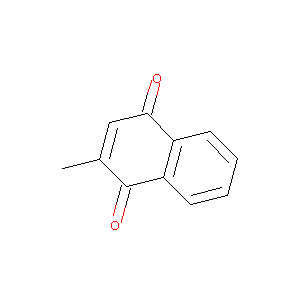 |
|||||||||||||||||||||||||||||||||||||||||||||
| 3D MOL | 2D MOL | |||||||||||||||||||||||||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 172.18 | ||||||||||||||||||||||||||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 2.2 | |||||||||||||||||||||||||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 0 | |||||||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 0 | |||||||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 2 | |||||||||||||||||||||||||||||||||||||||||||||
| ADMET Property | ||||||||||||||||||||||||||||||||||||||||||||||
| Adverse Drug Reaction (ADR) |
|
|||||||||||||||||||||||||||||||||||||||||||||
| Chemical Identifiers |
|
|||||||||||||||||||||||||||||||||||||||||||||
| Cross-matching ID | ||||||||||||||||||||||||||||||||||||||||||||||
| Combinatorial Drugs (CBD) | Click to Jump to the Detailed CBD Information of This Drug | |||||||||||||||||||||||||||||||||||||||||||||
| Repurposed Drugs (RPD) | Click to Jump to the Detailed RPD Information of This Drug | |||||||||||||||||||||||||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Transporter (DTP) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
