Details of the Drug
General Information of Drug (ID: DMUGQIV)
| Drug Name |
2-chloro-5-nitro-N-phenylbenzamide
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
2-Chloro-5-nitro-N-phenylbenzamide; GW9662; 22978-25-2; GW 9662; 2-Chloro-5-nitrobenzanilide; GW-9662; MLS001056751; CHEBI:79993; 2-Chloro-5-nitro-N-phenyl-benzamide; 2-Chloro-5-nitro-N-4-phenylbenzamide; benzamide, 2-chloro-5-nitro-N-phenyl-; SMR000326735; (2-chloro-5-nitrophenyl)-N-benzamide; SR-01000075999; Tocris-1508; Spectrum5_001989; Lopac-M-6191; AC1LD8S0; DSSTox_RID_79570; DSSTox_CID_20723; DSSTox_GSID_40723; Lopac0_000798; KBioGR_000361; KBioSS_000361; BSPBio_001021; SCHEMBL420231; CHEMBL375270; GTPL3442; cid_644213
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
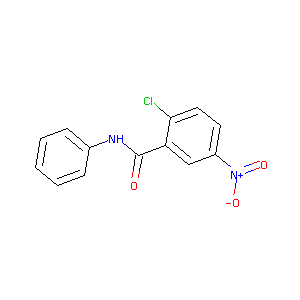 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 276.67 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 3.3 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 2 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 1 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 3 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| The Studied Disease | Discovery agent | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICD Disease Classification | N.A. | |||||||||||||||||||||||
|
||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||
References
