Details of the Drug
General Information of Drug (ID: DMYXELH)
| Drug Name |
1,2-NAPHTHOQUINONE
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
1,2-NAPHTHOQUINONE; 524-42-5; 1,2-Naphthalenedione; naphthalene-1,2-dione; o-Naphthoquinone; beta-Naphthoquinone; 1,2-Naphthaquinone; 1,2-Naftochinon; .beta.-Naphthoquinone; 1,2-Naftochinon [Czech]; CCRIS 1558; UNII-804K62F61Q; HSDB 2036; 1,2-dihydronaphthalene-1,2-dione; NSC 9831; EINECS 208-360-2; 1,2-Dihydro-1,2-diketo-naphthalene; BRN 0606546; CHEMBL52347; MLS000069467; AI3-14930; CHEBI:34055; C10H6O2; KETQAJRQOHHATG-UHFFFAOYSA-N; 804K62F61Q; SMR000059112; 1,2-Naphthoquinone, 95%, tech.; NAPHTHALENEDIONE; b-naphthoquinone
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
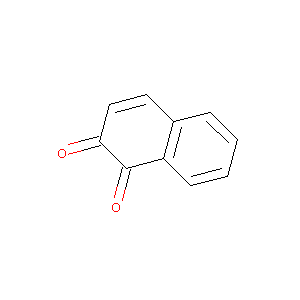 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 158.15 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 1.5 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 0 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 0 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 2 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| The Studied Disease | Discovery agent | |||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICD Disease Classification | N.A. | |||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||||||||||||||||||||
References
