Details of the Drug
General Information of Drug (ID: DMZC90M)
| Drug Name |
PD98059
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
167869-21-8; PD 98059; 2-(2-amino-3-methoxyphenyl)-4H-chromen-4-one; PD-98059; PD 98,059; 2-(2-amino-3-methoxyphenyl)chromen-4-one; 2-(2-Amino-3-methoxyphenyl)-4H-1-benzopyran-4-one; 2'-AMINO-3'-METHOXYFLAVONE; PD-098059; 4H-1-Benzopyran-4-one, 2-(2-Amino-3-methoxyphenyl)-; UNII-SJE1IO5E3I; SJE1IO5E3I; CHEMBL35482; C16H13NO3; CHEBI:77954; MFCD00671789; 2-(2-Amino-3-methoxy-phenyl)-chromen-4-one; SR-01000076097; PD 098059; 2-(2-Amino-3-methoxyphenyl)-chromen-4-one; NSC 679829; Tocris-1213; PD098059; Lopac-P-215; BiomolKI_000001; 2-(2-amino-3-methoxy-phenyl)chromen-4-one; BiomolKI2_000011; PD 98,059, solid; Lopac0_001028; BMK1-B1; BSPBio_000996; KBioGR_000336; KBioSS_000336; 2-(2'-amino-3'-methoxyphenyl)oxanaphthalen-4-one; cc-461; MLS006010134; SCHEMBL157826; 4H-1-Benzopyran-4-one,2-(2-amino-3-methoxyphenyl)-; GTPL5241; QCR-14; 2''-amino-3''-methoxyflavone; BCBcMAP01_000049; KBio2_000336; KBio2_002904; KBio2_005472; KBio3_000671; KBio3_000672; AOB2598; DTXSID40168416; BCPP000123; Bio1_000475; Bio1_000964; Bio1_001453; Bio2_000338; Bio2_000818; HMS1362B17; HMS1792B17; HMS1990B17; HMS3229M08; HMS3263M17; HMS3267D03; HMS3403B17; HMS3412O09; HMS3649N14; HMS3654I16; HMS3676O09; BCP02423; EX-A2127; ZINC1420826; Tox21_501028; ABP000927; BDBM50108771; NSC679828; s1177; AKOS015995212; BCP9001060; CCG-100605; CS-0169; LP01028; NSC 679828; NSC-679828; SB16629; SDCCGSBI-0051000.P003; IDI1_002093; SMP2_000052; NCGC00015790-01; NCGC00015790-02; NCGC00015790-03; NCGC00015790-04; NCGC00015790-05; NCGC00015790-06; NCGC00015790-07; NCGC00015790-08; NCGC00025045-01; NCGC00025045-02; NCGC00025045-03; NCGC00025045-04; NCGC00025045-05; NCGC00179347-01; NCGC00261713-01; AC-28412; AK-60664; AS-19374; HY-12028; NCI60_028554; SMR001456459; AB0033706; EU-0101028; FT-0716482; P-215; PD-98059/PD98059/; SW218254-2; X7398; EC-000.2425; A25454; P-4313; PD 98059 & Z-100; InSolution PD 98059 - CAS 167869-21-8; J-505513; SR-01000076097-1; SR-01000076097-3; SR-01000076097-6; 2-(2-amino-3-methoxyphenyl)-4-oxo-4H-[1]benzopyran; BRD-K62810658-001-05-6; BRD-K62810658-001-06-4; Q27088281; 2-(2-Amino-3-methoxy-phenyl)-chromen-4-one(PD98059); PD 98059 - CAS 167869-21-8; 4H-1-Benzopyran-4-one, 2-(2-amino-3-methoxyphenyl)- & Z-100
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
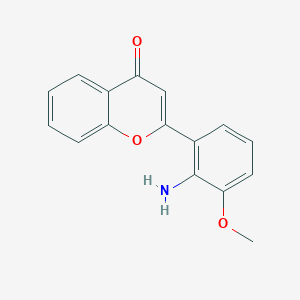 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 267.28 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 2.9 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 2 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 1 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 4 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| The Studied Disease | Cardiac arrest | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICD Disease Classification | MC82 | |||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||||||||
References
