| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
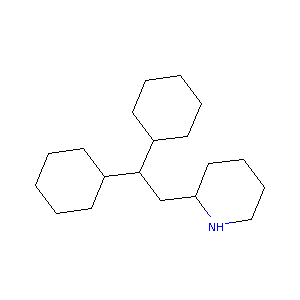
Perhexiline. Cardiovasc Drug Rev. 2007 Spring;25(1):76-97.
|
| 3 |
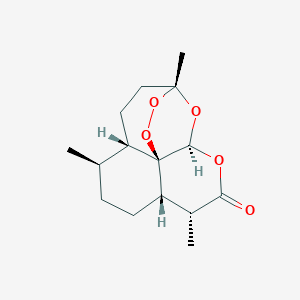
The fight against drug-resistant malaria: novel plasmodial targets and antimalarial drugs. Curr Med Chem. 2008;15(2):161-71.
|
| 4 |
A toxicogenomic approach to drug-induced phospholipidosis: analysis of its induction mechanism and establishment of a novel in vitro screening system. Toxicol Sci. 2005 Feb;83(2):282-92.
|
| 5 |
CYP2B6, CYP2D6, and CYP3A4 catalyze the primary oxidative metabolism of perhexiline enantiomers by human liver microsomes. Drug Metab Dispos. 2007 Jan;35(1):128-38.
|
| 6 |
Customised in vitro model to detect human metabolism-dependent idiosyncratic drug-induced liver injury. Arch Toxicol. 2018 Jan;92(1):383-399. doi: 10.1007/s00204-017-2036-4. Epub 2017 Jul 31.
|
| 7 |
Determination of phospholipidosis potential based on gene expression analysis in HepG2 cells. Toxicol Sci. 2007 Mar;96(1):101-14.
|
| 8 |
Effect of common medications on the expression of SARS-CoV-2 entry receptors in liver tissue. Arch Toxicol. 2020 Dec;94(12):4037-4041. doi: 10.1007/s00204-020-02869-1. Epub 2020 Aug 17.
|
| 9 |
Evaluating the Role of Multidrug Resistance Protein 3 (MDR3) Inhibition in Predicting Drug-Induced Liver Injury Using 125 Pharmaceuticals. Chem Res Toxicol. 2017 May 15;30(5):1219-1229. doi: 10.1021/acs.chemrestox.7b00048. Epub 2017 May 4.
|
| 10 |
Drug binding to the inactivated state is necessary but not sufficient for high-affinity binding to human ether--go-go-related gene channels. Mol Pharmacol. 2008 Nov;74(5):1443-52. doi: 10.1124/mol.108.049056. Epub 2008 Aug 13.
|
| 11 |
Antimalarial artemisinin drugs induce cytochrome P450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2005 Jun;67(6):1954-65.
|
| 12 |
Identification of the human cytochrome P450 enzymes involved in the in vitro metabolism of artemisinin. Br J Clin Pharmacol. 1999 Oct;48(4):528-35.
|
| 13 |
Inhibition of glutathione S-transferases by antimalarial drugs possible implications for circumventing anticancer drug resistance. Int J Cancer. 2002 Feb 10;97(5):700-5.
|
| 14 |
Antimalarial artemisinin drugs induce cytochrome P450 and MDR1 expression by activation of xenosensors pregnane X receptor and constitutive androstane receptor. Mol Pharmacol. 2005 Jun;67(6):1954-65.
|
| 15 |
Application of higher throughput screening (HTS) inhibition assays to evaluate the interaction of antiparasitic drugs with cytochrome P450s. Drug Metab Dispos. 2001 Jan;29(1):30-5.
|
| 16 |
Identification of Compounds That Inhibit Estrogen-Related Receptor Alpha Signaling Using High-Throughput Screening Assays. Molecules. 2019 Feb 27;24(5):841. doi: 10.3390/molecules24050841.
|
| 17 |
In vivo and mechanistic evidence of nuclear receptor CAR induction by artemisinin. Eur J Clin Invest. 2006 Sep;36(9):647-53. doi: 10.1111/j.1365-2362.2006.01700.x.
|
|
|
|
|
|
|