| 1 |
ClinicalTrials.gov (NCT04338698) Hydroxychloroquine, Oseltamivir and Azithromycin for the Treatment of COVID-19 Infection: An RCT
|
| 2 |
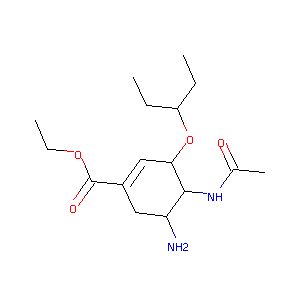
Oseltamivir FDA Label
|
| 3 |
Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007 Mar;70(3):461-77.
|
| 4 |
ClinicalTrials.gov (NCT04261270) A Randomized,Open,Controlled Clinical Study to Evaluate the Efficacy of ASC09F and Ritonavir for 2019-nCoV Pneumonia
|
| 5 |
Hydroxychloroquine FDA Label
|
| 6 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7198).
|
| 7 |
ClinicalTrials.gov (NCT04341727) Hydroxychloroquine,Hydroxychloroquine,Azithromycin in the Treatment of SARS CoV-2 Infection (WU352). U.S. National Institutes of Health.
|
| 8 |
Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 2008 Apr;78(1):91-102.
|
| 9 |
Nonclinical pharmacokinetics of oseltamivir and oseltamivir carboxylate in the central nervous system. Antimicrob Agents Chemother. 2009 Nov;53(11):4753-61.
|
| 10 |
Limited brain distribution of [3R,4R,5S]-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate phosphate (Ro 64-0802), a pharmacologically active form of oseltamivir, by active efflux across the blood-brain barrier mediated by organic anion transporter 3 (Oat3/Slc22a8) and multidrug resistance-associated protein 4 (Mrp4/Abcc4). Drug Metab Dispos. 2009 Feb;37(2):315-21.
|
| 11 |
Oseltamivir (tamiflu) is a substrate of peptide transporter 1. Drug Metab Dispos. 2009 Aug;37(8):1676-81.
|
| 12 |
FDA Drug Development and Drug Interactions
|
| 13 |
An in vitro coculture system of human peripheral blood mononuclear cells with hepatocellular carcinoma-derived cells for predicting drug-induced liver injury. Arch Toxicol. 2021 Jan;95(1):149-168. doi: 10.1007/s00204-020-02882-4. Epub 2020 Aug 20.
|
| 14 |
In vitro inhibition of carboxylesterase 1 by Kava (Piper methysticum) Kavalactones. Chem Biol Interact. 2022 Apr 25;357:109883. doi: 10.1016/j.cbi.2022.109883. Epub 2022 Mar 9.
|
| 15 |
Interleukin-6 alters the cellular responsiveness to clopidogrel, irinotecan, and oseltamivir by suppressing the expression of carboxylesterases HCE1 and HCE2. Mol Pharmacol. 2007 Sep;72(3):686-94. doi: 10.1124/mol.107.036889. Epub 2007 May 30.
|
| 16 |
Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 Mar;30(3):269-271.
|
| 17 |
TLR7/9 antagonists as therapeutics for immune-mediated inflammatory disorders. Inflamm Allergy Drug Targets. 2007 Dec;6(4):223-35.
|
| 18 |
Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat Rev Rheumatol. 2020 Mar;16(3):155-166.
|
| 19 |
MDR-ABC transporters: biomarkers in rheumatoid arthritis. Clin Exp Rheumatol. 2013 Sep-Oct;31(5):779-87.
|
| 20 |
Hydroxychloroquine: a physiologically-based pharmacokinetic model in the context of cancer-related autophagy modulation. J Pharmacol Exp Ther. 2018 Jun;365(3):447-459.
|
| 21 |
Somer M, Kallio J, Pesonen U, Pyykko K, Huupponen R, Scheinin M "Influence of hydroxychloroquine on the bioavailability of oral metoprolol." Br J Clin Pharmacol 49 (2000): 549-54. [PMID: 10848718]
|
| 22 |
Hydroxychloroquine-inhibited dengue virus is associated with host defense machinery. J Interferon Cytokine Res. 2015 Mar;35(3):143-56. doi: 10.1089/jir.2014.0038. Epub 2014 Oct 16.
|
| 23 |
Hydroxychloroquine potentiates Fas-mediated apoptosis of rheumatoid synoviocytes. Clin Exp Immunol. 2006 Jun;144(3):503-11. doi: 10.1111/j.1365-2249.2006.03070.x.
|
| 24 |
Hydroxychloroquine inhibits CD154 expression in CD4(+) T lymphocytes of systemic lupus erythematosus through NFAT, but not STAT5, signaling. Arthritis Res Ther. 2017 Aug 9;19(1):183. doi: 10.1186/s13075-017-1393-y.
|
| 25 |
Chloroquine and Hydroxychloroquine Increase Retinal Pigment Epithelial Layer Permeability. J Biochem Mol Toxicol. 2015 Jul;29(7):299-304. doi: 10.1002/jbt.21696. Epub 2015 Mar 9.
|
| 26 |
Hydroxychloroquine decreases human MSC-derived osteoblast differentiation and mineralization in vitro. J Cell Mol Med. 2018 Feb;22(2):873-882. doi: 10.1111/jcmm.13373. Epub 2017 Oct 3.
|
| 27 |
Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene. 2003 Jun 19;22(25):3927-36. doi: 10.1038/sj.onc.1206622.
|
| 28 |
Attenuation of chloroquine and hydroxychloroquine on the invasive potential of bladder cancer through targeting matrix metalloproteinase 2 expression. Environ Toxicol. 2021 Nov;36(11):2138-2145. doi: 10.1002/tox.23328. Epub 2021 Jul 19.
|
| 29 |
Reactive oxygen species mediate chloroquine-induced expression of chemokines by human astroglial cells. Glia. 2004 Jul;47(1):9-20. doi: 10.1002/glia.20017.
|
| 30 |
d-galactose induces premature senescence of lens epithelial cells by disturbing autophagy flux and mitochondrial functions. Toxicol Lett. 2018 Jun 1;289:99-106. doi: 10.1016/j.toxlet.2018.02.001. Epub 2018 Feb 6.
|
| 31 |
Antagonist-mediated down-regulation of Toll-like receptors increases the prevalence of human papillomavirus infection in systemic lupus erythematosus. Arthritis Res Ther. 2012 Apr 18;14(2):R80. doi: 10.1186/ar3803.
|
| 32 |
ClinicalTrials.gov (NCT04303299) Various Combination of Protease Inhibitors, Oseltamivir, Favipiravir, and Hydroxychloroquine for Treatment of COVID-19 : A Randomized Control Trial
|
|
|
|
|
|
|