Details of the Drug
General Information of Drug (ID: DMLPICK)
| Drug Name |
Quinidine
|
||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
Quinact; Quinaglute; Quinalan; Quinatime; Quindine; Quinicardine; Quinidex; Quinidine sulfate; Quiniduran; Quinora; beta-Quinine; Cardioquin; Chinidin; Chinidine; Cin-Quin; Cinchonan-9-ol, 6'-methoxy-, (9S)-; Conchinin; Conchinine; Conquinine; Duraquin; ITX08688JL; Kinidin; Pitayine; chinidinum; quinidina; quinidine; (+)-Quinidine; (8R,9S)-Quinidine; (9S)-6'-Methoxycinchonan-9-ol; (S)-[(2R,4S,5R)-5-ethenyl-1-azabicyclo[2.2.2]octan-2-yl](6-methoxyquinolin-4-yl)methanol; 56-54-2; CHEBI:28593; CHEMBL1294; MFCD00135581; UNII-ITX08688JL
|
||||||||||||||||||||||||||||||||||||||||
| Affected Organisms |
Humans and other mammals
|
||||||||||||||||||||||||||||||||||||||||
| ATC Code | |||||||||||||||||||||||||||||||||||||||||
| Structure |
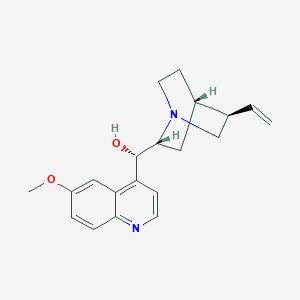 |
||||||||||||||||||||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 324.4 | |||||||||||||||||||||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 2.9 | ||||||||||||||||||||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 4 | ||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 1 | ||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 4 | ||||||||||||||||||||||||||||||||||||||||
| ADMET Property |
|
||||||||||||||||||||||||||||||||||||||||
| Adverse Drug Reaction (ADR) |
|
||||||||||||||||||||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||||||||||||||||||||
| Combinatorial Drugs (CBD) | Click to Jump to the Detailed CBD Information of This Drug | ||||||||||||||||||||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
