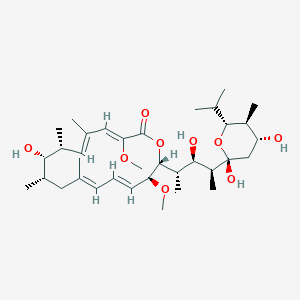
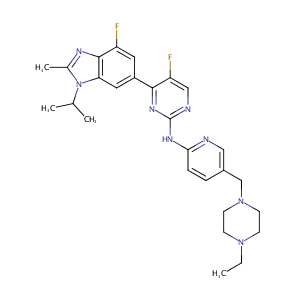
| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica. 2015 Mar;100(3):345-56.
|
| 3 |
2017 FDA drug approvals.Nat Rev Drug Discov. 2018 Feb;17(2):81-85.
|
| 4 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7382).
|
| 5 |
Autophagy mediates bronchial cell malignant transformation induced by chronic arsenic exposure via MEK/ERK1/2 pathway. Toxicol Lett. 2020 Oct 10;332:155-163. doi: 10.1016/j.toxlet.2020.06.006. Epub 2020 Jul 6.
|
| 6 |
Molecular basis of V-ATPase inhibition by bafilomycin A1. Nat Commun. 2021 Mar 19;12(1):1782.
|
| 7 |
Nano-sized iron particles may induce multiple pathways of cell death following generation of mistranscripted RNA in human corneal epithelial cells. Toxicol In Vitro. 2017 Aug;42:348-357.
|
| 8 |
Autophagy as a compensation mechanism participates in ethanol-induced fetal adrenal dysfunction in female rats. Toxicol Appl Pharmacol. 2018 Apr 15;345:36-47.
|
| 9 |
Vitamin D protects against particles-caused lung injury through induction of autophagy in an Nrf2-dependent manner. Environ Toxicol. 2019 May;34(5):594-609.
|
| 10 |
Chronic high glucose inhibits albumin reabsorption by lysosomal alkalinization in cultured porcine proximal tubular epithelial cells (LLC-PK1). Diabetes Res Clin Pract. 2006 Jun;72(3):223-30. doi: 10.1016/j.diabres.2005.10.019. Epub 2005 Nov 28.
|
| 11 |
Berbamine Hydrochloride inhibits lysosomal acidification by activating Nox2 to potentiate chemotherapy-induced apoptosis via the ROS-MAPK pathway in human lung carcinoma cells. Cell Biol Toxicol. 2023 Aug;39(4):1297-1317. doi: 10.1007/s10565-022-09756-8. Epub 2022 Sep 7.
|
| 12 |
SB202190 inhibits endothelial cell apoptosis via induction of autophagy and heme oxygenase-1. Oncotarget. 2018 May 1;9(33):23149-23163. doi: 10.18632/oncotarget.25234. eCollection 2018 May 1.
|
| 13 |
5-Methoxyflavanone induces cell cycle arrest at the G2/M phase, apoptosis and autophagy in HCT116 human colon cancer cells. Toxicol Appl Pharmacol. 2011 Aug 1;254(3):288-98. doi: 10.1016/j.taap.2011.05.003. Epub 2011 May 15.
|
| 14 |
GJA1 reverses arsenic-induced EMT via modulating MAPK/ERK signaling pathway. Toxicol Appl Pharmacol. 2022 Sep 1;450:116138. doi: 10.1016/j.taap.2022.116138. Epub 2022 Jun 21.
|
| 15 |
Combined effects of EGFR tyrosine kinase inhibitors and vATPase inhibitors in NSCLC cells. Toxicol Appl Pharmacol. 2015 Aug 15;287(1):17-25. doi: 10.1016/j.taap.2015.05.001. Epub 2015 May 14.
|
| 16 |
Autophagy induction by capsaicin in malignant human breast cells is modulated by p38 and extracellular signal-regulated mitogen-activated protein kinases and retards cell death by suppressing endoplasmic reticulum stress-mediated apoptosis. Mol Pharmacol. 2010 Jul;78(1):114-25. doi: 10.1124/mol.110.063495. Epub 2010 Apr 6.
|
| 17 |
Cadmium induces triglyceride levels via microsomal triglyceride transfer protein (MTTP) accumulation caused by lysosomal deacidification regulated by endoplasmic reticulum (ER) Ca(2+) homeostasis. Chem Biol Interact. 2021 Oct 1;348:109649. doi: 10.1016/j.cbi.2021.109649. Epub 2021 Sep 10.
|
| 18 |
Inhibition of cholesterol metabolism underlies synergy between mTOR pathway inhibition and chloroquine in bladder cancer cells. Oncogene. 2016 Aug 25;35(34):4518-28. doi: 10.1038/onc.2015.511. Epub 2016 Feb 8.
|
| 19 |
Resveratrol-activated AMPK/SIRT1/autophagy in cellular models of Parkinson's disease. Neurosignals. 2011;19(3):163-74. doi: 10.1159/000328516. Epub 2011 Jul 22.
|
| 20 |
LncRNA UCA1 attenuates autophagy-dependent cell death through blocking autophagic flux under arsenic stress. Toxicol Lett. 2018 Mar 1;284:195-204. doi: 10.1016/j.toxlet.2017.12.009. Epub 2017 Dec 15.
|
| 21 |
Transcriptomic Landscape of Cisplatin-Resistant Neuroblastoma Cells. Cells. 2019 Mar 12;8(3):235. doi: 10.3390/cells8030235.
|
| 22 |
G2019S LRRK2 mutant fibroblasts from Parkinson's disease patients show increased sensitivity to neurotoxin 1-methyl-4-phenylpyridinium dependent of autophagy. Toxicology. 2014 Oct 3;324:1-9. doi: 10.1016/j.tox.2014.07.001. Epub 2014 Jul 10.
|
| 23 |
Biological specificity of CDK4/6 inhibitors: dose response relationship, in vivo signaling, and composite response signature. Oncotarget. 2017 Jul 4;8(27):43678-43691. doi: 10.18632/oncotarget.18435.
|
| 24 |
Interpreting expression profiles of cancers by genome-wide survey of breadth of expression in normal tissues. Genomics 2005 Aug;86(2):127-41.
|
| 25 |
Abemaciclib Inhibits Renal Tubular Secretion Without Changing Glomerular Filtration Rate. Clin Pharmacol Ther. 2019 May;105(5):1187-1195.
|
| 26 |
LiverTox: Clinical and Research Information on Drug-Induced Liver Injury [Internet]-Abemaciclib.
|
| 27 |
CDK4/6 Inhibition Controls Proliferation of Bladder Cancer and Transcription of RB1. J Urol. 2016 Mar;195(3):771-9. doi: 10.1016/j.juro.2015.08.082. Epub 2015 Aug 28.
|
| 28 |
Effect of abemaciclib (LY2835219) on enhancement of chemotherapeutic agents in ABCB1 and ABCG2 overexpressing cells in vitro and in vivo. Biochem Pharmacol. 2017 Jan 15;124:29-42. doi: 10.1016/j.bcp.2016.10.015. Epub 2016 Nov 2.
|
| 29 |
In vitro therapeutic effects of abemaciclib on triple-negative breast cancer cells. J Biochem Mol Toxicol. 2021 Sep;35(9):e22858. doi: 10.1002/jbt.22858. Epub 2021 Jul 26.
|
|
|
|
|
|
|