Details of the Drug
General Information of Drug (ID: DMNBA0G)
| Drug Name |
BENZOQUINONE
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
p-benzoquinone; Benzoquinone; 1,4-BENZOQUINONE; Quinone; 106-51-4; p-Quinone; Chinone; 2,5-Cyclohexadiene-1,4-dione; para-Benzoquinone; cyclohexa-2,5-diene-1,4-dione; Cyclohexadienedione; para-Quinone; 1,4-Benzoquine; Steara pbq; 1,4-Dioxybenzene; p-Chinon; 1,4-Cyclohexadienedione; Benzo-chinon; Benzo-1,4-quinone; 1,4-Diossibenzene; Chinon; 1,4-Dioxy-benzol; 1,4-Cyclohexadiene dioxide; Semiquinone anion; semiquinone radicals; RCRA waste number U197; p-Chinon [German]; NCI-C55845; Caswell No. 719C; Benzo-chinon [German]; USAF P-220
|
||||||||||||||||||||||
| Indication |
|
||||||||||||||||||||||
| ATC Code | |||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
||||||||||||||||||||||
| Structure |
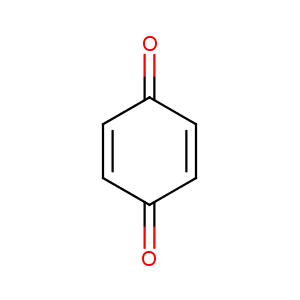 |
||||||||||||||||||||||
| 3D MOL | 2D MOL | ||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 108.09 | |||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 0.2 | ||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 0 | ||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 0 | ||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 2 | ||||||||||||||||||||||
| Chemical Identifiers |
|
||||||||||||||||||||||
| Cross-matching ID | |||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| The Studied Disease | Discovery agent | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICD Disease Classification | N.A. | |||||||||||||||||||||||
|
||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||
References
