| 1 |
ClinicalTrials.gov (NCT01867138) Neonatal Suspected Sepsis Treated With Cefazolin or Vancomycin
|
| 2 |
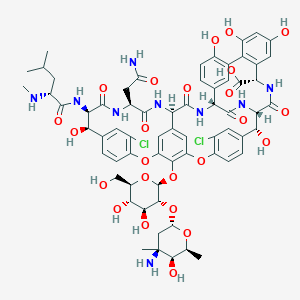
Vancomycin FDA Label
|
| 3 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015
|
| 4 |
The ChEMBL database in 2017. Nucleic Acids Res. 2017 Jan 4;45(D1):D945-D954.
|
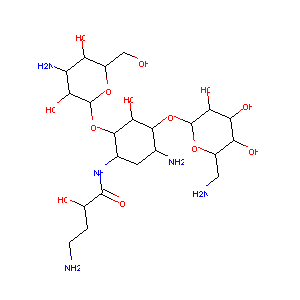
| 5 |
Amikacin FDA Label
|
| 6 |
Emerging drugs for bacterial urinary tract infections. Expert Opin Emerg Drugs. 2005 May;10(2):275-98.
|
| 7 |
Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993 Aug;37(8):1563-71.
|
| 8 |
Sequence of the vanC gene of Enterococcus gallinarum BM4174 encoding a D-alanine:D-alanine ligase-related protein necessary for vancomycin resistance. Gene. 1992 Mar 1;112(1):53-8.
|
| 9 |
vanG element insertions within a conserved chromosomal site conferring vancomycin resistance to Streptococcus agalactiae and Streptococcus anginosus. mBio. 2014 Jul 22;5(4):e01386-14.
|
| 10 |
Bacterial resistance to vancomycin: overproduction, purification, and characterization of VanC2 from Enterococcus casseliflavus as a D-Ala-D-Ser ligase. Proc Natl Acad Sci U S A. 1997 Sep 16;94(19):10040-4.
|
| 11 |
vanI: a novel D-Ala-D-Lac vancomycin resistance gene cluster found in Desulfitobacterium hafniense. Microb Biotechnol. 2014 Sep;7(5):456-66.
|
| 12 |
Emergence of high-level resistance to glycopeptides in Enterococcus gallinarum and Enterococcus casseliflavus. Antimicrob Agents Chemother. 1994 Jul;38(7):1675-7.
|
| 13 |
vanA in Enterococcus faecium, Enterococcus faecalis, and Enterococcus casseliflavus detected in French cattle. Foodborne Pathog Dis. 2009 Nov;6(9):1107-11.
|
| 14 |
Systems pharmacological analysis of drugs inducing stevens-johnson syndrome and toxic epidermal necrolysis. Chem Res Toxicol. 2015 May 18;28(5):927-34. doi: 10.1021/tx5005248. Epub 2015 Apr 3.
|
| 15 |
[Successful treatment of MRSA-associated glomerulonephritis with antibiotic therapy]. Nihon Jinzo Gakkai Shi. 2003;45(1):37-41.
|
| 16 |
Modulation of melanogenesis and antioxidant defense system in melanocytes by amikacin. Toxicol In Vitro. 2013 Apr;27(3):1102-8.
|
| 17 |
Bacterial resistance to aminoglycosides and beta-lactams: the Tn1331 transposon paradigm. Front Biosci. 2000 Jan 1;5:D20-9.
|
| 18 |
Relationship between antimicrobial resistance and aminoglycoside-modifying enzyme gene expressions in Acinetobacter baumannii. Chin Med J (Engl). 2005 Jan 20;118(2):141-5.
|
| 19 |
Expression of Clostridium thermocellum endoglucanase gene in Lactobacillus gasseri and Lactobacillus johnsonii and characterization of the genetically modified probiotic lactobacilli. Curr Microbiol. 2000 Apr;40(4):257-63.
|
| 20 |
An in vitro coculture system of human peripheral blood mononuclear cells with hepatocellular carcinoma-derived cells for predicting drug-induced liver injury. Arch Toxicol. 2021 Jan;95(1):149-168. doi: 10.1007/s00204-020-02882-4. Epub 2020 Aug 20.
|
| 21 |
Amikacin-induced type 5 Bartter-like syndrome with severe hypocalcemia. J Postgrad Med. 2009 Jul-Sep;55(3):208-10. doi: 10.4103/0022-3859.57407.
|
| 22 |
Translational readthrough by the aminoglycoside geneticin (G418) modulates SMN stability in vitro and improves motor function in SMA mice in vivo. Hum Mol Genet. 2009 Apr 1;18(7):1310-22. doi: 10.1093/hmg/ddp030. Epub 2009 Jan 15.
|
| 23 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
|
|
|
|
|
|