| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
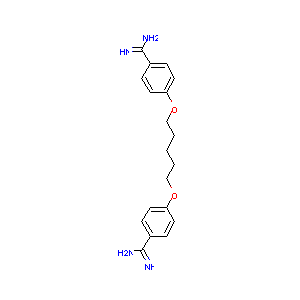
Pentamidine FDA Label
|
| 3 |
Opportunities and challenges in antiparasitic drug discovery. Nat Rev Drug Discov. 2005 Sep;4(9):727-40.
|
| 4 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015
|
| 5 |
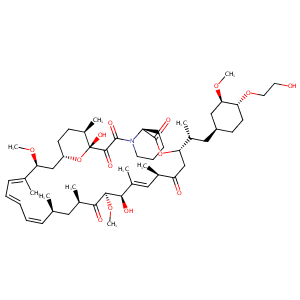
Everolimus FDA Label
|
| 6 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5889).
|
| 7 |
Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016 May;15(5):327-47.
|
| 8 |
A toxicogenomic approach to drug-induced phospholipidosis: analysis of its induction mechanism and establishment of a novel in vitro screening system. Toxicol Sci. 2005 Feb;83(2):282-92.
|
| 9 |
Inhibitors of tryptase for the treatment of mast cell-mediated diseases. Curr Pharm Des. 1998 Oct;4(5):381-96.
|
| 10 |
Transport of dicationic drugs pentamidine and furamidine by human organic cation transporters. Drug Metab Dispos. 2009 Feb;37(2):424-30.
|
| 11 |
Identification of human cytochrome P(450)s that metabolise anti-parasitic drugs and predictions of in vivo drug hepatic clearance from in vitro data. Eur J Clin Pharmacol. 2003 Sep;59(5-6):429-42.
|
| 12 |
Value of preemptive CYP2C19 genotyping in allogeneic stem cell transplant patients considered for pentamidine administration. Clin Transplant. 2011 May-Jun;25(3):E271-5.
|
| 13 |
Application of higher throughput screening (HTS) inhibition assays to evaluate the interaction of antiparasitic drugs with cytochrome P450s. Drug Metab Dispos. 2001 Jan;29(1):30-5.
|
| 14 |
Bisbenzamidine derivative, pentamidine represses DNA damage response through inhibition of histone H2A acetylation. Mol Cancer. 2010 Feb 9;9:34. doi: 10.1186/1476-4598-9-34.
|
| 15 |
Profiling the immunotoxicity of chemicals based on in vitro evaluation by a combination of the Multi-ImmunoTox assay and the IL-8 Luc assay. Arch Toxicol. 2018 Jun;92(6):2043-2054. doi: 10.1007/s00204-018-2199-7. Epub 2018 Mar 29.
|
| 16 |
Systems pharmacological analysis of drugs inducing stevens-johnson syndrome and toxic epidermal necrolysis. Chem Res Toxicol. 2015 May 18;28(5):927-34. doi: 10.1021/tx5005248. Epub 2015 Apr 3.
|
| 17 |
Antileishmanial drugs cause up-regulation of interferon-gamma receptor 1, not only in the monocytes of visceral leishmaniasis cases but also in cultured THP1 cells. Ann Trop Med Parasitol. 2003 Apr;97(3):245-57. doi: 10.1179/000349803235001714.
|
| 18 |
Identification of human Ether--go-go related gene modulators by three screening platforms in an academic drug-discovery setting. Assay Drug Dev Technol. 2010 Dec;8(6):727-42. doi: 10.1089/adt.2010.0331.
|
| 19 |
Association of CYP1A1 and CYP1B1 inhibition in in vitro assays with drug-induced liver injury. J Toxicol Sci. 2021;46(4):167-176. doi: 10.2131/jts.46.167.
|
| 20 |
Investigation of imatinib and other approved drugs as starting points for antidiabetic drug discovery with FXR modulating activity. Biochem Pharmacol. 2012 Jun 15;83(12):1674-81. doi: 10.1016/j.bcp.2012.02.027. Epub 2012 Mar 7.
|
| 21 |
Mammalian target of rapamycin, its mode of action and clinical response in metastatic clear cell carcinoma. Gan To Kagaku Ryoho. 2009 Jul;36(7):1076-9.
|
| 22 |
Closer to the Site of Action: Everolimus Concentrations in Peripheral Blood Mononuclear Cells Correlate Well With Whole Blood Concentrations. Ther Drug Monit. 2015 Oct;37(5):675-80.
|
| 23 |
The evolving experience using everolimus in clinical transplantation. Transplant Proc. 2004 Mar;36(2 Suppl):495S-499S.
|
|
|
|
|
|
|