Details of the Drug
General Information of Drug (ID: DM3BH1Y)
| Drug Name |
Lapatinib
|
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
FMM; Tycerb; Lapatinib Ditosylate; Lapatinib [INN]; Lapatinib tosilate hydrate; GSK 572016; GSK572016; GW 572016; GW 572016X; GW572016; Lapatinib (INN); Tykerb (TN); Lapatinib, Tykerb, GW572016; N-[3-chloro-4-[(3-fluorophenyl)methoxy]phenyl]-6-[5-[(2-methylsulfonylethylamino)methyl]furan-2-yl]quinazolin-4-amine; N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE; N-(3-Chloro-4-((3-fluorophenyl)methoxy)phenyl)-6-(5-((2-methylsulfonylethylamino)methyl)-2-furyl)quinazolin-4-amine; N-(3-Chloro-4-{[(3-fluorophenyl)methyl]oxy}phenyl)-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furanyl]-4-quinazolinamine; 4-[[3-Chloro-4-(3-fluorobenzyloxy)phenyl]amino]-6-[5-[[(2-methanesulfonylethyl)amino]methyl]furan-2-yl]quinazoline; Lapatinib (ERBB2 inhibitor)
|
|||||||||||||||||||||||||||||||||||
| Indication |
|
|||||||||||||||||||||||||||||||||||
| Therapeutic Class |
Anticancer Agents
|
|||||||||||||||||||||||||||||||||||
| Affected Organisms |
Humans and other mammals
|
|||||||||||||||||||||||||||||||||||
| ATC Code | ||||||||||||||||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
|||||||||||||||||||||||||||||||||||
| Structure |
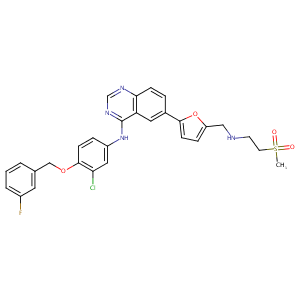 |
|||||||||||||||||||||||||||||||||||
| 3D MOL | 2D MOL | |||||||||||||||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 3 | Molecular Weight (mw) | 581.1 | ||||||||||||||||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 5.1 | |||||||||||||||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 11 | |||||||||||||||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 2 | |||||||||||||||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 9 | |||||||||||||||||||||||||||||||||||
| ADMET Property |
|
|||||||||||||||||||||||||||||||||||
| Adverse Drug Reaction (ADR) |
|
|||||||||||||||||||||||||||||||||||
| Chemical Identifiers |
|
|||||||||||||||||||||||||||||||||||
| Cross-matching ID | ||||||||||||||||||||||||||||||||||||
| Combinatorial Drugs (CBD) | Click to Jump to the Detailed CBD Information of This Drug | |||||||||||||||||||||||||||||||||||
| Repurposed Drugs (RPD) | Click to Jump to the Detailed RPD Information of This Drug | |||||||||||||||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Transporter (DTP) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Expression Atlas of This Drug
| The Studied Disease | Breast cancer | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICD Disease Classification | 2C60-2C65 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Expression Atlas (MEA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Experimental Cancer Drug Sensitivity Information
Drug-Drug Interaction (DDI) Information of This Drug
|
Coadministration of a Drug Treating the Same Disease as Lapatinib
Coadministration of a Drug Treating the Disease Different from Lapatinib (Comorbidity)
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Drug Inactive Ingredient(s) (DIG) and Formulation(s) of This Drug
| DIG |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pharmaceutical Formulation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
| 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5692). | ||||
|---|---|---|---|---|---|
| 2 | Lapatinib FDA Label | ||||
| 3 | BDDCS applied to over 900 drugs | ||||
| 4 | Trend Analysis of a Database of Intravenous Pharmacokinetic Parameters in Humans for 1352 Drug Compounds | ||||
| 5 | Estimating the safe starting dose in phase I clinical trials and no observed effect level based on QSAR modeling of the human maximum recommended daily dose | ||||
| 6 | HLA-DQA1*02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J Clin Oncol. 2011 Feb 20;29(6):667-73. doi: 10.1200/JCO.2010.31.3197. Epub 2011 Jan 18. | ||||
| 7 | Triple negative breast cancer--current status and prospective targeted treatment based on HER1 (EGFR), TOP2A and C-MYC gene assessment. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2009 Mar;153(1):13-7. | ||||
| 8 | Inhibition of eEF-2 kinase sensitizes human nasopharyngeal carcinoma cells to lapatinib-induced apoptosis through the Src and Erk pathways.BMC Cancer. 2016 Oct 19;16(1):813. | ||||
| 9 | The role of efflux and uptake transporters in [N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine (GW572016, lapatinib) disposition and drug interactions. Drug Metab Dispos. 2008 Apr;36(4):695-701. | ||||
| 10 | Tarascon Pocket Pharmacopoeia 2018 Classic Shirt-Pocket Edition. | ||||
| 11 | Mechanism-based inactivation of cytochrome P450 3A4 by lapatinib. Mol Pharmacol. 2010 Oct;78(4):693-703. | ||||
| 12 | Clinical pharmacokinetics of tyrosine kinase inhibitors. Cancer Treat Rev. 2009 Dec;35(8):692-706. | ||||
| 13 | Cytotoxicity of 34 FDA approved small-molecule kinase inhibitors in primary rat and human hepatocytes. Toxicol Lett. 2018 Jul;291:138-148. doi: 10.1016/j.toxlet.2018.04.010. Epub 2018 Apr 12. | ||||
| 14 | Effects of lapatinib on cell proliferation and apoptosis in NB4 cells. Oncol Lett. 2018 Jan;15(1):235-242. doi: 10.3892/ol.2017.7342. Epub 2017 Nov 3. | ||||
| 15 | Combining lapatinib (GW572016), a small molecule inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing breast cancer cells. Oncogene. 2005 Sep 15;24(41):6213-21. doi: 10.1038/sj.onc.1208774. | ||||
| 16 | Interference with bile salt export pump function is a susceptibility factor for human liver injury in drug development. Toxicol Sci. 2010 Dec; 118(2):485-500. | ||||
| 17 | The involvement of hepatic cytochrome P450s in the cytotoxicity of lapatinib. Toxicol Sci. 2023 Dec 21;197(1):69-78. doi: 10.1093/toxsci/kfad099. | ||||
| 18 | CDK4/6 inhibition provides a potent adjunct to Her2-targeted therapies in preclinical breast cancer models. Genes Cancer. 2014 Jul;5(7-8):261-72. doi: 10.18632/genesandcancer.24. | ||||
| 19 | The dual ErbB1/ErbB2 inhibitor, lapatinib (GW572016), cooperates with tamoxifen to inhibit both cell proliferation- and estrogen-dependent gene expression in antiestrogen-resistant breast cancer. Cancer Res. 2005 Jan 1;65(1):18-25. | ||||
| 20 | Canadian Pharmacists Association. | ||||
| 21 | Cerner Multum, Inc. "UK Summary of Product Characteristics.". | ||||
| 22 | Product Information. Ixempra (ixabepilone). Bristol-Myers Squibb, Princeton, NJ. | ||||
| 23 | Product Information. Verzenio (abemaciclib). Lilly, Eli and Company, Indianapolis, IN. | ||||
| 24 | Product Information. Tykerb (lapatinib). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 25 | Cerner Multum, Inc. "Australian Product Information.". | ||||
| 26 | Product Information. Jevtana (cabazitaxel). sanofi-aventis , Bridgewater, NJ. | ||||
| 27 | Product Information. Tibsovo (ivosidenib). Agios Pharmaceuticals, Cambridge, MA. | ||||
| 28 | Product Information. Sirturo (bedaquiline). Janssen Pharmaceuticals, Titusville, NJ. | ||||
| 29 | Product Information. Arcapta Neohaler (indacaterol). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 30 | Bengtsson B, Fagerstrom PO "Extrapulmonary effects of terbutaline during prolonged administration." Clin Pharmacol Ther 31 (1982): 726-32. [PMID: 7042176] | ||||
| 31 | Product Information. Balversa (erdafitinib). Janssen Products, LP, Horsham, PA. | ||||
| 32 | Product Information. Turalio (pexidartinib). Daiichi Sankyo, Inc., Parsippany, NJ. | ||||
| 33 | Product Information. Opsumit (macitentan). Actelion Pharmaceuticals US Inc, South San Francisco, CA. | ||||
| 34 | Product Information. Daurismo (glasdegib). Pfizer U.S. Pharmaceuticals Group, New York, NY. | ||||
| 35 | Product Information. Nexletol (bempedoic acid). Esperion Therapeutics, Ann Arbor, MI. | ||||
| 36 | Product Information. Fycompa (perampanel). Eisai Inc, Teaneck, NJ. | ||||
| 37 | Product Information. Ofev (nintedanib). Boehringer Ingelheim, Ridgefield, CT. | ||||
| 38 | Product Information. Kalydeco (ivacaftor). Vertex Pharmaceuticals, Cambridge, MA. | ||||
| 39 | Product Information. Xarelto (rivaroxaban). Bayer Inc, Toronto, IA. | ||||
| 40 | Product Information. Polivy (polatuzumab vedotin). Genentech, South San Francisco, CA. | ||||
| 41 | Product Information. Austedo (deutetrabenazine). Teva Pharmaceuticals USA, North Wales, PA. | ||||
| 42 | Product Information. Ingrezza (valbenazine). Neurocrine Biosciences, Inc., San Diego, CA. | ||||
| 43 | Product Information. Aliqopa (copanlisib). Bayer Pharmaceutical Inc, West Haven, CT. | ||||
| 44 | Product Information. VESIcare (solifenacin). GlaxoSmithKline, Research Triangle Park, NC. | ||||
| 45 | Product Information. Myrbetriq (mirabegron). Astellas Pharma US, Inc, Deerfield, IL. | ||||
| 46 | Product Information. Harvoni (ledipasvir-sofosbuvir). Gilead Sciences, Foster City, CA. | ||||
| 47 | Product Information. Vosevi (sofosbuvir/velpatasvir/voxilaprevir). Gilead Sciences, Foster City, CA. | ||||
| 48 | Product Information. VFEND (voriconazole). Pfizer U.S. Pharmaceuticals, New York, NY. | ||||
| 49 | Product Information. Pifeltro (doravirine). Merck & Company Inc, Whitehouse Station, NJ. | ||||
| 50 | Product Information. Rukobia (fostemsavir). ViiV Healthcare, Research Triangle Park, NC. | ||||
| 51 | Product Information. Kynamro (mipomersen). Genzyme Corporation, Cambridge, MA. | ||||
| 52 | Product Information. Juxtapid (lomitapide). Aegerion Pharmaceuticals Inc, Cambridge, MA. | ||||
| 53 | Product Information. Samsca (tolvaptan). Otsuka American Pharmaceuticals Inc, Rockville, MD. | ||||
| 54 | Product Information. Orladeyo (berotralstat). BioCryst Pharmaceuticals Inc, Durham, NC. | ||||
| 55 | Product Information. Caplyta (lumateperone). Intra-Cellular Therapies, Inc., New York, NY. | ||||
| 56 | Product Information. Movantik (naloxegol). Astra-Zeneca Pharmaceuticals, Wilmington, DE. | ||||
| 57 | Product Information. Xalkori (crizotinib). Pfizer U.S. Pharmaceuticals Group, New York, NY. | ||||
| 58 | Product Information. Zepzelca (lurbinectedin). Jazz Pharmaceuticals, Palo Alto, CA. | ||||
| 59 | Product Information. Tagrisso (osimertinib). Astra-Zeneca Pharmaceuticals, Wilmington, DE. | ||||
| 60 | Product Information. Gavreto (pralsetinib). Blueprint Medicines Corporation, Cambridge, MA. | ||||
| 61 | Product Information. Tabrecta (capmatinib). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 62 | Harper KM, Knapp DJ, Criswell HE, Breese GR "Vasopressin and alcohol: A multifaceted relationship." Psychopharmacology (Berl) 235 (2018): 3363-79. [PMID: 32936259] | ||||
| 63 | Al-Nawakil C, Willems L, Mauprivez C, et.al "Successful treatment of l-asparaginase-induced severe acute hepatotoxicity using mitochondrial cofactors." Leuk Lymphoma 55 (2014): 1670-4. [PMID: 24090500] | ||||
| 64 | Product Information. Calquence (acalabrutinib). Astra-Zeneca Pharmaceuticals, Wilmington, DE. | ||||
| 65 | Product Information. Imbruvica (ibrutinib). Pharmacyclics Inc, Sunnyvale, CA. | ||||
| 66 | Product Information. Koselugo (selumetinib). Astra-Zeneca Pharmaceuticals, Wilmington, DE. | ||||
| 67 | Product Information. Braftovi (encorafenib). Array BioPharma Inc., Boulder, CO. | ||||
| 68 | Product Information. Ubrelvy (ubrogepant). Allergan Inc, Irvine, CA. | ||||
| 69 | Product Information. Nurtec ODT (rimegepant). Biohaven Pharmaceuticals, New Haven, CT. | ||||
| 70 | Product Information. Reyvow (lasmiditan). Lilly, Eli and Company, Indianapolis, IN. | ||||
| 71 | Product Information. Exjade (deferasirox). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 72 | Product Information. Addyi (flibanserin). Sprout Pharmaceuticals, Raleigh, NC. | ||||
| 73 | Product Information. Farydak (panobinostat). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 74 | Product Information. Zeposia (ozanimod). Celgene Corporation, Summit, NJ. | ||||
| 75 | Product Information. Varubi (rolapitant). Tesaro Inc., Waltham, MA. | ||||
| 76 | Product Information. Lynparza (olaparib). Astra-Zeneca Pharmaceuticals, Wilmington, DE. | ||||
| 77 | Product Information. Nuplazid (pimavanserin). Accelis Pharma, East Windsor, NJ. | ||||
| 78 | Product Information. Xeglyze (abametapir topical). Dr. Reddy's Laboratories Inc, Upper Saddle River, NJ. | ||||
| 79 | Product Information. Macrilen (macimorelin). Aeterna Zentaris, Charleston, SC. | ||||
| 80 | Product Information. Xenleta (lefamulin). Nabriva Therapeutics US, Inc., King of Prussia, PA. | ||||
| 81 | Product Information. Orgovyx (relugolix). Myovant Sciences, Inc., Brisbane, CA. | ||||
| 82 | Product Information. Nubeqa (darolutamide). Bayer HealthCare Pharmaceuticals Inc., Whippany, NJ. | ||||
| 83 | Product Information. Rapaflo (silodosin). Watson Pharmaceuticals, Corona, CA. | ||||
| 84 | Product Information. Uptravi (selexipag). Actelion Pharmaceuticals US Inc, South San Francisco, CA. | ||||
| 85 | Product Information. Afinitor (everolimus). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 86 | Product Information. Torisel (temsirolimus). Wyeth-Ayerst Laboratories, Philadelphia, PA. | ||||
| 87 | Product Information. Barhemsys (amisulpride). Acacia Pharma, Inc, Indianapolis, IN. | ||||
| 88 | Product Information. Odomzo (sonidegib). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 89 | Product Information. Tavalisse (fostamatinib). Rigel Pharmaceuticals, South San Francisco, CA. | ||||
| 90 | EMA. European Medicines Agency. European Union "EMA - List of medicines under additional monitoring.". | ||||
| 91 | Allred AJ, Bowen CJ, Park JW, et al. "Eltrombopag increases plasma rosuvastatin exposure in healthy volunteers." Br J Clin Pharmacol 72 (2011): 321-9. [PMID: 21434975] | ||||
