| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
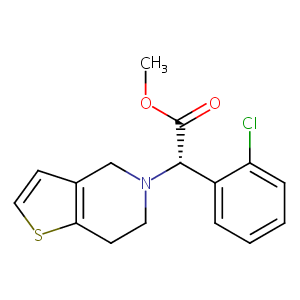
Clopidogrel FDA Label
|
| 3 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7150).
|
| 4 |
Preventing Cardiac Complication of COVID-19 Disease With Early Acute Coronary Syndrome Therapy: A Randomised Controlled Trial. (C-19-ACS)
|
| 5 |
Allopurinol FDA Label
|
| 6 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 6795).
|
| 7 |
P2Y12, a new platelet ADP receptor, target of clopidogrel. Semin Vasc Med. 2003 May;3(2):113-22.
|
| 8 |
Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther. 2006 Nov;80(5):486-501.
|
| 9 |
Substrates, inducers, inhibitors and structure-activity relationships of human Cytochrome P450 2C9 and implications in drug development. Curr Med Chem. 2009;16(27):3480-675.
|
| 10 |
Clinical pharmacokinetics and pharmacodynamics of clopidogrel. Clin Pharmacokinet. 2015 Feb;54(2):147-66.
|
| 11 |
Cytochrome P450 3A inhibition by ketoconazole affects prasugrel and clopidogrel pharmacokinetics and pharmacodynamics differently. Clin Pharmacol Ther. 2007 May;81(5):735-41.
|
| 12 |
Clopidogrel pathway. Pharmacogenet Genomics. 2010 Jul;20(7):463-5.
|
| 13 |
Impact of the CYP2C19 gene polymorphism on clopidogrel personalized drug regimen and the clinical outcomes. Clin Lab. 2016 Sep 1;62(9):1773-1780.
|
| 14 |
Identification of novel agonists by high-throughput screening and molecular modelling of human constitutive androstane receptor isoform 3. Arch Toxicol. 2019 Aug;93(8):2247-2264. doi: 10.1007/s00204-019-02495-6. Epub 2019 Jul 16.
|
| 15 |
Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010 Jan;38(1):92-9. doi: 10.1124/dmd.109.029132.
|
| 16 |
Increased risk of atherothrombotic events associated with cytochrome P450 3A5 polymorphism in patients taking clopidogrel. CMAJ. 2006 Jun 6;174(12):1715-22. doi: 10.1503/cmaj.060664.
|
| 17 |
Platelet reactivity and clopidogrel resistance are associated with the H2 haplotype of the P2Y12-ADP receptor gene. Int J Cardiol. 2009 Apr 17;133(3):341-5. doi: 10.1016/j.ijcard.2007.12.118. Epub 2008 May 15.
|
| 18 |
Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013 Sep;94(3):317-23. doi: 10.1038/clpt.2013.105. Epub 2013 May 22.
|
| 19 |
Angiogenesis inhibitor SR 25989 upregulates thrombospondin-1 expression in human vascular endothelial cells and foreskin fibroblasts. Biol Cell. 1997 Jul;89(4):295-307.
|
| 20 |
Association of CYP1A1 and CYP1B1 inhibition in in vitro assays with drug-induced liver injury. J Toxicol Sci. 2021;46(4):167-176. doi: 10.2131/jts.46.167.
|
| 21 |
Clopidogrel increases expression of chemokines in peripheral blood mononuclear cells in patients with coronary artery disease: results of a double-blind placebo-controlled study. J Thromb Haemost. 2006 Oct;4(10):2140-7. doi: 10.1111/j.1538-7836.2006.02131.x. Epub 2006 Jul 17.
|
| 22 |
Differential effect of clopidogrel and aspirin on the release of BDNF from platelets. J Neuroimmunol. 2011 Sep 15;238(1-2):104-6. doi: 10.1016/j.jneuroim.2011.06.015. Epub 2011 Jul 31.
|
| 23 |
Attenuated expression of the tight junction proteins is involved in clopidogrel-induced gastric injury through p38 MAPK activation. Toxicology. 2013 Feb 8;304:41-8. doi: 10.1016/j.tox.2012.11.020. Epub 2012 Dec 7.
|
| 24 |
PAR-1 genotype influences platelet aggregation and procoagulant responses in patients with coronary artery disease prior to and during clopidogrel therapy. Platelets. 2005 Sep;16(6):340-5. doi: 10.1080/00207230500120294.
|
| 25 |
Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med. 2011 Jan;17(1):110-6. doi: 10.1038/nm.2281. Epub 2010 Dec 19.
|
| 26 |
Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007 Dec;5(12):2429-36. doi: 10.1111/j.1538-7836.2007.02775.x. Epub 2007 Sep 26.
|
| 27 |
Interleukin-6 alters the cellular responsiveness to clopidogrel, irinotecan, and oseltamivir by suppressing the expression of carboxylesterases HCE1 and HCE2. Mol Pharmacol. 2007 Sep;72(3):686-94. doi: 10.1124/mol.107.036889. Epub 2007 May 30.
|
| 28 |
High loading dose of clopidogrel is unable to satisfactorily inhibit platelet reactivity in patients with glycoprotein IIIA gene polymorphism: a genetic substudy of PRAGUE-8 trial. Blood Coagul Fibrinolysis. 2009 Jun;20(4):257-62. doi: 10.1097/mbc.0b013e328325455b.
|
| 29 |
Uric acid-lowering treatment with benzbromarone in patients with heart failure: a double-blind placebo-controlled crossover preliminary study. Circ Heart Fail. 2010 Jan;3(1):73-81.
|
| 30 |
Allopurinol: xanthine oxidase inhibitor. Tex Med. 1966 Jan;62(1):100-1.
|
| 31 |
Isolation, characterization and differential gene expression of multispecific organic anion transporter 2 in mice. Mol Pharmacol. 2002 Jul;62(1):7-14.
|
| 32 |
Renal transport of organic compounds mediated by mouse organic anion transporter 3 (mOat3): further substrate specificity of mOat3. Drug Metab Dispos. 2004 May;32(5):479-83.
|
| 33 |
Xanthine oxidase inhibition by allopurinol affects the reliability of urinary caffeine metabolic ratios as markers for N-acetyltransferase 2 and CYP1A2 activities. Eur J Clin Pharmacol. 1999 Jan;54(11):869-76.
|
| 34 |
Transport mechanism and substrate specificity of human organic anion transporter 2 (hOat2 [SLC22A7]). J Pharm Pharmacol. 2005 May;57(5):573-8.
|
| 35 |
HLA-B*5801 allele as a genetic marker for severe cutaneous adverse reactions caused by allopurinol. Proc Natl Acad Sci U S A. 2005 Mar 15;102(11):4134-9. doi: 10.1073/pnas.0409500102. Epub 2005 Mar 2.
|
| 36 |
A study of HLA class I and class II 4-digit allele level in Stevens-Johnson syndrome and toxic epidermal necrolysis. Int J Immunogenet. 2011 Aug;38(4):303-9. doi: 10.1111/j.1744-313X.2011.01011.x. Epub 2011 May 4.
|
| 37 |
Positive and negative associations of HLA class I alleles with allopurinol-induced SCARs in Koreans. Pharmacogenet Genomics. 2011 May;21(5):303-7. doi: 10.1097/FPC.0b013e32834282b8.
|
| 38 |
Effect of common medications on the expression of SARS-CoV-2 entry receptors in liver tissue. Arch Toxicol. 2020 Dec;94(12):4037-4041. doi: 10.1007/s00204-020-02869-1. Epub 2020 Aug 17.
|
| 39 |
Drug-induced hepatic steatosis in absence of severe mitochondrial dysfunction in HepaRG cells: proof of multiple mechanism-based toxicity. Cell Biol Toxicol. 2021 Apr;37(2):151-175. doi: 10.1007/s10565-020-09537-1. Epub 2020 Jun 14.
|
| 40 |
Selection of drugs to test the specificity of the Tg.AC assay by screening for induction of the gadd153 promoter in vitro. Toxicol Sci. 2003 Aug;74(2):260-70. doi: 10.1093/toxsci/kfg113. Epub 2003 May 2.
|
| 41 |
Allopurinol induces innate immune responses through mitogen-activated protein kinase signaling pathways in HL-60 cells. J Appl Toxicol. 2016 Sep;36(9):1120-8. doi: 10.1002/jat.3272. Epub 2015 Dec 7.
|
| 42 |
Systemic drugs inducing non-immediate cutaneous adverse reactions and contact sensitizers evoke similar responses in THP-1 cells. J Appl Toxicol. 2015 Apr;35(4):398-406. doi: 10.1002/jat.3033. Epub 2014 Aug 4.
|
| 43 |
Allopurinol Protects Against Cholestatic Liver Injury in Mice Not Through Depletion of Uric Acid. Toxicol Sci. 2021 May 27;181(2):295-305. doi: 10.1093/toxsci/kfab034.
|
| 44 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
| 45 |
Clinical Pharmacogenetics Implementation Consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clin Pharmacol Ther. 2013 Feb;93(2):153-8. doi: 10.1038/clpt.2012.209. Epub 2012 Oct 17.
|
|
|
|
|
|
|