Details of the Drug
General Information of Drug (ID: DM4O2Z7)
| Drug Name |
Cyclophosphamide
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synonyms |
ASTA; Ciclofosfamida; Ciclophosphamide; Clafen; Claphene; Cycloblastin; Cyclophosphamid; Cyclophosphamides; Cyclophosphamidum; Cyclophosphan; Cyclophosphane; Cyclophosphanum; Cyclophosphoramide; Cyclostin; Cyklofosfamid; Cytophosphan; Cytophosphane; Cytoxan; Endoxan; Endoxana; Endoxanal; Endoxane; Enduxan; Genoxal; Mitoxan; Neosar; Procytox; Revimmune; Semdoxan; Sendoxan; Senduxan; Zyklophosphamid; Ciclophosphamide [INN]; Cyclophosphamide Sterile; Cyclophosphamide anhydrous; Cyklofosfamid [Czech]; Cytoxan Lyoph; Endoxan R; Lyophilized Cytoxan; Zyklophosphamid [German]; ASTA B518; Asta B 518; B 518; C 0768; CB 4564; SK 20501; B-518; CB-4564; Ciclofosfamida [INN-Spanish]; Cyclophosphamide (INN); Cyclophosphamide (TN); Cyclophosphamide (anhydrous form); Cyclophosphamide (anhydrous); Cyclophosphamidum [INN-Latin]; Cytoxan (TN); Endoxan (TN); Endoxan-Asta; Neosar (TN); Occupation, cyclophosphamide exposure; Procytox (TN); Revimmune (TN); Bis(2-Chloroethyl)phosphami de cyclic propanolamide; Bis(2-Chloroethyl)phosphamide cyclic propanolamide ester; Bis(2-chloroethyl)phosphoramide cyclic propanolamide ester; D,L-Cyclophosphamide; Cyclophosphamide, (+-)-Isomer; N,N-Bis(2-chloroethyl)-1,3,2-oxazaphosphinan-2-amine 2-oxide; (+-)-Cyclophosphamide; (-)-Cyclophosphamide; (RS)-Cyclophosphamide; 1-(bis(2-chloroethyl)amino)-1-oxo-2-aza-5-oxaphosphoridine; 1-Bis(2-chloroethyl)amino-1-oxo-2-aza-5-oxaphosphoridin; 4-Hydroxy-cyclophosphan-mamophosphatide
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Indication |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Therapeutic Class |
Anticancer Agents
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Affected Organisms |
Humans and other mammals
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ATC Code | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Drug Type |
Small molecular drug
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure |
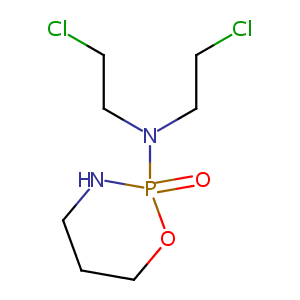 |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3D MOL | 2D MOL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| #Ro5 Violations (Lipinski): 0 | Molecular Weight (mw) | 261.079 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Logarithm of the Partition Coefficient (xlogp) | 0.6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Rotatable Bond Count (rotbonds) | 5 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Donor Count (hbonddonor) | 1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Hydrogen Bond Acceptor Count (hbondacc) | 4 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ADMET Property |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Adverse Drug Reaction (ADR) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Identifiers |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cross-matching ID | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Combinatorial Drugs (CBD) | Click to Jump to the Detailed CBD Information of This Drug | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Repurposed Drugs (RPD) | Click to Jump to the Detailed RPD Information of This Drug | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Molecular Interaction Atlas of This Drug
 Drug Therapeutic Target (DTT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 Drug Transporter (DTP) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug-Metabolizing Enzyme (DME) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 Drug Off-Target (DOT) |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Interaction Atlas (MIA) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Experimental Cancer Drug Sensitivity Information
Drug-Drug Interaction (DDI) Information of This Drug
|
Coadministration of a Drug Treating the Same Disease as Cyclophosphamide
Coadministration of a Drug Treating the Disease Different from Cyclophosphamide (Comorbidity)
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Drug Inactive Ingredient(s) (DIG) and Formulation(s) of This Drug
| DIG |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pharmaceutical Formulation |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
| 1 | Cyclophosphamide FDA Label | ||||
|---|---|---|---|---|---|
| 2 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7154). | ||||
| 3 | BDDCS applied to over 900 drugs | ||||
| 4 | Critical Evaluation of Human Oral Bioavailability for Pharmaceutical Drugs by Using Various Cheminformatics Approaches | ||||
| 5 | Trend Analysis of a Database of Intravenous Pharmacokinetic Parameters in Humans for 1352 Drug Compounds | ||||
| 6 | Estimating the safe starting dose in phase I clinical trials and no observed effect level based on QSAR modeling of the human maximum recommended daily dose | ||||
| 7 | ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899. | ||||
| 8 | O6-methylguanine-DNA methyltransferase activity and sensitivity to cyclophosphamide and cisplatin in human lung tumor xenografts. Int J Cancer. 1998 Sep 11;77(6):919-22. | ||||
| 9 | Interaction of oxazaphosphorines with multidrug resistance-associated protein 4 (MRP4). AAPS J. 2010 Sep;12(3):300-8. | ||||
| 10 | Effects of ketoconazole on cyclophosphamide metabolism: evaluation of CYP3A4 inhibition effect using the in vitro and in vivo models. Exp Anim. 2018 Feb 9;67(1):71-82. | ||||
| 11 | CYP2C9 polymorphisms in human tumors. Anticancer Res. 2006 Jan-Feb;26(1A):299-305. | ||||
| 12 | Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006 Mar 13;25(11):1679-91. | ||||
| 13 | Development of a substrate-activity based approach to identify the major human liver P-450 catalysts of cyclophosphamide and ifosfamide activation based on cDNA-expressed activities and liver microsomal P-450 profiles. Drug Metab Dispos. 1999 Jun;27(6):655-66. | ||||
| 14 | The effect of cyclophosphamide with and without dexamethasone on cytochrome P450 3A4 and 2B6 in human hepatocytes. Drug Metab Dispos. 2002 Jul;30(7):814-22. | ||||
| 15 | Enhanced cyclophosphamide and ifosfamide activation in primary human hepatocyte cultures: response to cytochrome P-450 inducers and autoinduction by oxazaphosphorines. Cancer Res. 1997 May 15;57(10):1946-54. | ||||
| 16 | Summary of information on human CYP enzymes: human P450 metabolism data. Drug Metab Rev. 2002 Feb-May;34(1-2):83-448. | ||||
| 17 | Genomic profiling uncovers a molecular pattern for toxicological characterization of mutagens and promutagens in vitro. Toxicol Sci. 2011 Jul;122(1):185-97. | ||||
| 18 | Effect of cyclophosphamide on gene expression of cytochromes p450 and beta-actin in the HL-60 cell line. Eur J Pharmacol. 2002 Aug 9;449(3):197-205. | ||||
| 19 | Transcriptome-based functional classifiers for direct immunotoxicity. Arch Toxicol. 2014 Mar;88(3):673-89. | ||||
| 20 | Comparative gene expression analysis of a chronic myelogenous leukemia cell line resistant to cyclophosphamide using oligonucleotide arrays and response to tyrosine kinase inhibitors. Leuk Res. 2007 Nov;31(11):1511-20. | ||||
| 21 | Relations between polymorphisms in drug-metabolising enzymes and toxicity of chemotherapy with cyclophosphamide, thiotepa and carboplatin. Pharmacogenet Genomics. 2008 Nov;18(11):1009-15. doi: 10.1097/FPC.0b013e328313aaa4. | ||||
| 22 | Effect of common medications on the expression of SARS-CoV-2 entry receptors in liver tissue. Arch Toxicol. 2020 Dec;94(12):4037-4041. doi: 10.1007/s00204-020-02869-1. Epub 2020 Aug 17. | ||||
| 23 | DSouza DL, Levasseur LM, Nezamis J, Robbins DK, Simms L, Koch KM "Effect of alosetron on the pharmacokinetics of alprazolam." J Clin Pharmacol 41 (2001): 452-4. [PMID: 11304902] | ||||
| 24 | Cerner Multum, Inc. "Australian Product Information.". | ||||
| 25 | Abad S, Moachon L, Blanche P, Bavoux F, Sicard D, Salmon-Ceron D "Possible interaction between glicazide, fluconazole and sulfamethoxazole resulting in severe hypoglycaemia." Br J Clin Pharmacol 52 (2001): 456-7. [PMID: 11678792] | ||||
| 26 | Cerner Multum, Inc. "UK Summary of Product Characteristics.". | ||||
| 27 | Johnson EJ, MacGowan AP, Potter MN, et al "Reduced absorption of oral ciprofloxacin after chemotherapy for haematological malignancy." J Antimicrob Chemother 25 (1990): 837-42. [PMID: 2373666] | ||||
| 28 | Product Information. Synercid (dalfopristin-quinupristin) Rhone-Poulenc Rorer, Collegeville, PA. | ||||
| 29 | Product Information. Piqray (alpelisib). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 30 | Multum Information Services, Inc. Expert Review Panel. | ||||
| 31 | Product Information. Prevymis (letermovir). Merck & Company Inc, Whitehouse Station, NJ. | ||||
| 32 | Charasson V, Haaz MC, Robert J "Determination of Drug Interactions Occurring with the Metabolic Pathways of Irinotecan." Drug Metab Dispos 30 (2002): 731-733. [PMID: 12019202] | ||||
| 33 | Product Information. Serzone (nefazodone). Bristol-Myers Squibb, Princeton, NJ. | ||||
| 34 | Product Information. Trileptal (oxcarbazepine) Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 35 | Product Information. Xcopri (cenobamate). SK Life Science, Inc., Paramus, NJ. | ||||
| 36 | EMEA. European Medicines Agency "EPARs. European Union Public Assessment Reports.". | ||||
| 37 | Garattini S, Spreafico F "Some examples of interactions between drugs in cancer chemotherapy." Antibiot Chemother 23 (1978): 283-94. [PMID: 348085] | ||||
| 38 | Chang TK, Yu L, Maurel P, Waxman DJ "Enhanced cyclophosphamide and ifosfamide activation in primary human hepatocyte cultures: response to cytochrome P-450 inducers and autoinduction by oxazaphosphorines." Cancer Res 57 (1997): 1946-54. [PMID: 9157990] | ||||
| 39 | Product Information. Agenerase (amprenavir). Glaxo Wellcome, Research Triangle Pk, NC. | ||||
| 40 | Product Information. Prezista (darunavir). Ortho Biotech Inc, Bridgewater, NJ. | ||||
| 41 | Product Information. Arava (leflunomide). Hoechst Marion-Roussel Inc, Kansas City, MO. | ||||
| 42 | Product Information. Orladeyo (berotralstat). BioCryst Pharmaceuticals Inc, Durham, NC. | ||||
| 43 | Product Information. Prolia (denosumab). Amgen USA, Thousand Oaks, CA. | ||||
| 44 | Product Information. Lorbrena (lorlatinib). Pfizer U.S. Pharmaceuticals Group, New York, NY. | ||||
| 45 | Product Information. Copiktra (duvelisib). Verastem, Inc., Needham, MA. | ||||
| 46 | Product Information. Clolar (clofarabine). sanofi-aventis, Bridgewater, NJ. | ||||
| 47 | Bennett CL, Nebeker JR, Samore MH, et al "The Research on Adverse Drug Events and Reports (RADAR) project." JAMA 293 (2005): 2131-40. [PMID: 15870417] | ||||
| 48 | Product Information. Vumerity (diroximel fumarate). Alkermes, Inc, Cambridge, MA. | ||||
| 49 | Product Information. Gilenya (fingolimod). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 50 | Product Information. Ocrevus (ocrelizumab). Genentech, South San Francisco, CA. | ||||
| 51 | Product Information. Gleevec (imatinib mesylate). Novartis Pharmaceuticals, East Hanover, NJ. | ||||
| 52 | Product Information. Synribo (omacetaxine). Teva Pharmaceuticals USA, North Wales, PA. | ||||
| 53 | Product Information. Provigil (modafinil). Cephalon, Inc, West Chester, PA. | ||||
| 54 | Agencia Espaola de Medicamentos y Productos Sanitarios Healthcare "Centro de informacion online de medicamentos de la AEMPS - CIMA.". | ||||
| 55 | EMA. European Medicines Agency. European Union "EMA - List of medicines under additional monitoring.". | ||||
| 56 | Product Information. Xeglyze (abametapir topical). Dr. Reddy's Laboratories Inc, Upper Saddle River, NJ. | ||||
| 57 | Product Information. Fycompa (perampanel). Eisai Inc, Teaneck, NJ. | ||||
| 58 | Benoist G, van Oort I, et al "Drug-drug interaction potential in men treated with enzalutamide: Mind the gap." Br J Clin Pharmacol 0 (2017): epub. [PMID: 28881501] | ||||
| 59 | Product Information. Tracleer (bosentan). Acetelion Pharmaceuticals US, Inc, South San Francisco, CA. | ||||
| 60 | Product Information. Prograf (tacrolimus). Fujisawa, Deerfield, IL. | ||||
| 61 | Product Information. Arcalyst (rilonacept). Regeneron Pharmaceuticals Inc, Tarrytown, NY. | ||||
| 62 | Product Information. Cimzia (certolizumab). UCB Pharma Inc, Smyrna, GA. | ||||
| 63 | CDC. Centers for Disease Control and Prevention/ "Recommendations of the advisory committtee on immunization practices (ACIP): use of vaccines and immune globulins in persons with altered immunocompetence." MMWR Morb Mortal Wkly Rep 42(RR-04) (1993): 1-18. [PMID: 20300058] | ||||
| 64 | Product Information. Tavalisse (fostamatinib). Rigel Pharmaceuticals, South San Francisco, CA. | ||||
| 65 | Shaunak S, Munro JM, Weinbren K, Walport MJ, Cox TM "Cyclophosphamide-induced liver necrosis: a possible interaction with azathioprine." Q J Med 67 (1988): 309-17. [PMID: 3060893] | ||||
