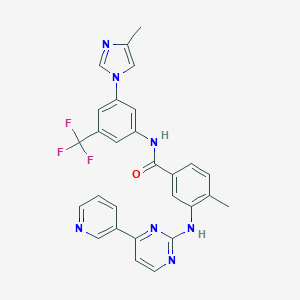
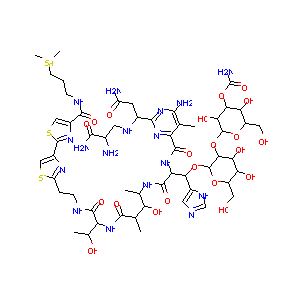
| 1 |
Loss of function mutations in VARS encoding cytoplasmic valyl-tRNA synthetase cause microcephaly, seizures, and progressive cerebral atrophy.Hum Genet. 2018 Apr;137(4):293-303. doi: 10.1007/s00439-018-1882-3. Epub 2018 Apr 24.
|
| 2 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5697).
|
| 3 |
Bleomycin FDA Label
|
| 4 |
Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007 Mar;70(3):461-77.
|
| 5 |
Assessment of the inhibition potential of Licochalcone A against human UDP-glucuronosyltransferases. Food Chem Toxicol. 2016 Apr;90:112-22.
|
| 6 |
Exposure-based assessment of chemical teratogenicity using morphogenetic aggregates of human embryonic stem cells. Reprod Toxicol. 2020 Jan;91:74-91. doi: 10.1016/j.reprotox.2019.10.004. Epub 2019 Nov 8.
|
| 7 |
Endoplasmic reticulum stress-mediated apoptosis in imatinib-resistant leukemic K562-r cells triggered by AMN107 combined with arsenic trioxide. Exp Biol Med (Maywood). 2013 Aug 1;238(8):932-42. doi: 10.1177/1535370213492689. Epub 2013 Jul 24.
|
| 8 |
2007 FDA drug approvals: a year of flux. Nat Rev Drug Discov. 2008 Feb;7(2):107-9.
|
| 9 |
Interaction of nilotinib, dasatinib and bosutinib with ABCB1 and ABCG2: implications for altered anti-cancer effects and pharmacological properties. Br J Pharmacol. 2009 Oct;158(4):1153-64.
|
| 10 |
KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017 Jan 4;45(D1):D353-D361. (dg:DG01665)
|
| 11 |
Contribution of OATP1B1 and OATP1B3 to the disposition of sorafenib and sorafenib-glucuronide. Clin Cancer Res. 2013 Mar 15;19(6):1458-66.
|
| 12 |
Drug interactions with the tyrosine kinase inhibitors imatinib, dasatinib, and nilotinib. Blood. 2011 Feb 24;117(8):e75-87.
|
| 13 |
Role of cytochrome P450 2C8 in drug metabolism and interactions. Pharmacol Rev. 2016 Jan;68(1):168-241.
|
| 14 |
Resistance to daunorubicin, imatinib, or nilotinib depends on expression levels of ABCB1 and ABCG2 in human leukemia cells. Chem Biol Interact. 2014 Aug 5;219:203-10. doi: 10.1016/j.cbi.2014.06.009. Epub 2014 Jun 19.
|
| 15 |
Reversal of ABCB1 mediated efflux by imatinib and nilotinib in cells expressing various transporter levels. Chem Biol Interact. 2017 Aug 1;273:171-179. doi: 10.1016/j.cbi.2017.06.012. Epub 2017 Jun 13.
|
| 16 |
Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes. Toxicol Appl Pharmacol. 2013 Oct 1;272(1):245-55.
|
| 17 |
p53 Gene (NY-CO-13) Levels in Patients with Chronic Myeloid Leukemia: The Role of Imatinib and Nilotinib. Diseases. 2018 Jan 25;6(1):13. doi: 10.3390/diseases6010013.
|
| 18 |
Nilotinib reduced the viability of human ovarian cancer cells via mitochondria-dependent apoptosis, independent of JNK activation. Toxicol In Vitro. 2016 Mar;31:1-11. doi: 10.1016/j.tiv.2015.11.002. Epub 2015 Nov 6.
|
| 19 |
AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009 Nov 6;16(5):401-12. doi: 10.1016/j.ccr.2009.09.028.
|
| 20 |
The CML-related oncoprotein BCR/ABL induces expression of histidine decarboxylase (HDC) and the synthesis of histamine in leukemic cells. Blood. 2006 Nov 15;108(10):3538-47. doi: 10.1182/blood-2005-12-028456. Epub 2006 Jul 18.
|
| 21 |
Cytotoxicity of 34 FDA approved small-molecule kinase inhibitors in primary rat and human hepatocytes. Toxicol Lett. 2018 Jul;291:138-148. doi: 10.1016/j.toxlet.2018.04.010. Epub 2018 Apr 12.
|
| 22 |
Development and validation of the TGx-HDACi transcriptomic biomarker to detect histone deacetylase inhibitors in human TK6 cells. Arch Toxicol. 2021 May;95(5):1631-1645. doi: 10.1007/s00204-021-03014-2. Epub 2021 Mar 26.
|
| 23 |
Pulmonary fibrosis model using micro-CT analyzable human PSC-derived alveolar organoids containing alveolar macrophage-like cells. Cell Biol Toxicol. 2022 Aug;38(4):557-575. doi: 10.1007/s10565-022-09698-1. Epub 2022 Mar 10.
|
| 24 |
An antisense oligonucleotide targeted to human Ku86 messenger RNA sensitizes M059K malignant glioma cells to ionizing radiation, bleomycin, and etoposide but not DNA cross-linking agents. Cancer Res. 2002 Oct 15;62(20):5888-96.
|
| 25 |
XRCC1 deficiency sensitizes human lung epithelial cells to genotoxicity by crocidolite asbestos and Libby amphibole. Environ Health Perspect. 2010 Dec;118(12):1707-13. doi: 10.1289/ehp.1002312. Epub 2010 Aug 11.
|
| 26 |
Bleomycin and talisomycin sequence-specific strand scission of DNA: a mechanism of double-strand cleavage. Cancer Res. 1982 Jul;42(7):2779-85.
|
| 27 |
The C-terminus of human bleomycin hydrolase is required for protection against bleomycin-induced chromosomal damage. Mutat Res. 1998 Oct 12;421(1):1-7.
|
| 28 |
Genetic polymorphisms of DNA repair and xenobiotic-metabolizing enzymes: role in mutagen sensitivity. Carcinogenesis. 2002 Jun;23(6):1003-8. doi: 10.1093/carcin/23.6.1003.
|
| 29 |
Age-dependent basal level and induction capacity of copper-zinc and manganese superoxide dismutase and other scavenging enzyme activities in leukocytes from young and elderly adults. Am J Pathol. 1993 Jul;143(1):312-20.
|
| 30 |
Agents associated with lung inflammation induce similar responses in NCI-H292 lung epithelial cells. Toxicol In Vitro. 2008 Oct;22(7):1782-8.
|
| 31 |
Characterization of DNA reactive and non-DNA reactive anticancer drugs by gene expression profiling. Mutat Res. 2007 Jun 1;619(1-2):16-29. doi: 10.1016/j.mrfmmm.2006.12.007. Epub 2007 Feb 8.
|
| 32 |
Direct activation of ATM by resveratrol under oxidizing conditions. PLoS One. 2014 Jun 16;9(6):e97969. doi: 10.1371/journal.pone.0097969. eCollection 2014.
|
| 33 |
Enhanced IL-1 beta and tumor necrosis factor-alpha release and messenger RNA expression in macrophages from idiopathic pulmonary fibrosis or after asbestos exposure. J Immunol. 1993 May 1;150(9):4188-96.
|
| 34 |
Synergistic anticancer activity of dietary tea polyphenols and bleomycin hydrochloride in human cervical cancer cell: Caspase-dependent and independent apoptotic pathways. Chem Biol Interact. 2016 Mar 5;247:1-10. doi: 10.1016/j.cbi.2016.01.012. Epub 2016 Jan 29.
|
| 35 |
Distinct mechanisms of cell-kill by triapine and its terminally dimethylated derivative Dp44mT due to a loss or gain of activity of their copper(II) complexes. Biochem Pharmacol. 2014 Oct 1;91(3):312-22. doi: 10.1016/j.bcp.2014.08.006. Epub 2014 Aug 15.
|
| 36 |
Radiation-induced cathepsin S is involved in radioresistance. Int J Cancer. 2009 Apr 15;124(8):1794-801. doi: 10.1002/ijc.24095.
|
| 37 |
Bleomycin-induced nuclear factor-kappaB activation in human bronchial epithelial cells involves the phosphorylation of glycogen synthase kinase 3beta. Toxicol Lett. 2009 Jun 22;187(3):194-200. doi: 10.1016/j.toxlet.2009.02.023. Epub 2009 Mar 13.
|
| 38 |
Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res. 2000 May;6(5):1796-803.
|
| 39 |
DNA methylation inhibitor 5-Aza-2'-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol Cell Biol. 2008 Jan;28(2):752-71. doi: 10.1128/MCB.01799-07. Epub 2007 Nov 8.
|
|
|
|
|
|
|