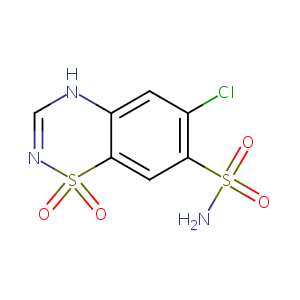
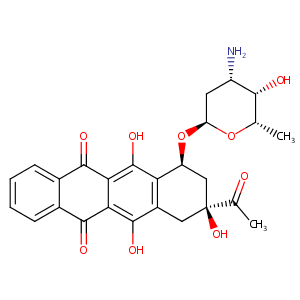
| DOT Name |
DOT ID |
UniProt ID |
Mode of Action |
REF |
|
Volume-regulated anion channel subunit LRRC8B (LRRC8B)
|
OTH2LMJ4
|
LRC8B_HUMAN
|
Increases ADR
|
[10] |
|
Growth arrest-specific protein 7 (GAS7)
|
OT0M5TNY
|
GAS7_HUMAN
|
Increases ADR
|
[10] |
|
LisH domain-containing protein ARMC9 (ARMC9)
|
OT0MZER2
|
ARMC9_HUMAN
|
Increases ADR
|
[6] |
|
E3 ubiquitin-protein ligase NEDD4-like (NEDD4L)
|
OT1B19RU
|
NED4L_HUMAN
|
Increases ADR
|
[11] |
|
Protein-lysine 6-oxidase (LOX)
|
OT1C2HIU
|
LYOX_HUMAN
|
Increases ADR
|
[6] |
|
Alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase (MGAT1)
|
OT1E09PZ
|
MGAT1_HUMAN
|
Increases ADR
|
[6] |
|
Adenylate cyclase type 9 (ADCY9)
|
OT1IZT5K
|
ADCY9_HUMAN
|
Increases ADR
|
[12] |
|
Beta-arrestin-1 (ARRB1)
|
OT2NJS37
|
ARRB1_HUMAN
|
Increases ADR
|
[6] |
|
Endoplasmic reticulum-Golgi intermediate compartment protein 1 (ERGIC1)
|
OT351FKB
|
ERGI1_HUMAN
|
Increases ADR
|
[10] |
|
Neutral ceramidase (ASAH2)
|
OT47TIF3
|
ASAH2_HUMAN
|
Increases ADR
|
[6] |
|
P2Y purinoceptor 2 (P2RY2)
|
OT47XN46
|
P2RY2_HUMAN
|
Increases ADR
|
[11] |
|
Protein phosphatase 1 regulatory subunit 12A (PPP1R12A)
|
OT4AVU95
|
MYPT1_HUMAN
|
Increases ADR
|
[6] |
|
Protein argonaute-2 (AGO2)
|
OT4JY32Q
|
AGO2_HUMAN
|
Increases ADR
|
[6] |
|
Glutamate--cysteine ligase regulatory subunit (GCLM)
|
OT6CP234
|
GSH0_HUMAN
|
Increases ADR
|
[6] |
|
Zinc transporter ZIP11 (SLC39A11)
|
OT71LCT2
|
S39AB_HUMAN
|
Increases ADR
|
[6] |
|
Triple functional domain protein (TRIO)
|
OT71X1AK
|
TRIO_HUMAN
|
Increases ADR
|
[13] |
|
Signaling threshold-regulating transmembrane adapter 1 (SIT1)
|
OT7MDF09
|
SIT1_HUMAN
|
Increases ADR
|
[10] |
|
Cytosolic carboxypeptidase 4 (AGBL1)
|
OT8NQLH0
|
CBPC4_HUMAN
|
Increases ADR
|
[6] |
|
2-oxoisovalerate dehydrogenase subunit beta, mitochondrial (BCKDHB)
|
OT8OSVYU
|
ODBB_HUMAN
|
Increases ADR
|
[6] |
|
Apolipoprotein L3 (APOL3)
|
OT95SQHR
|
APOL3_HUMAN
|
Increases ADR
|
[10] |
|
Proline-rich membrane anchor 1 (PRIMA1)
|
OT9ITT3P
|
PRIMA_HUMAN
|
Increases ADR
|
[11] |
|
CD44 antigen (CD44)
|
OT9TTJ41
|
CD44_HUMAN
|
Increases ADR
|
[6] |
|
Golgi integral membrane protein 4 (GOLIM4)
|
OTB5NV8A
|
GOLI4_HUMAN
|
Increases ADR
|
[6] |
|
Protein Jade-1 (JADE1)
|
OTBPJIRI
|
JADE1_HUMAN
|
Increases ADR
|
[6] |
|
Trinucleotide repeat-containing gene 6C protein (TNRC6C)
|
OTBR07PM
|
TNR6C_HUMAN
|
Increases ADR
|
[6] |
|
NAD-capped RNA hydrolase NUDT12 (NUDT12)
|
OTBUY58C
|
NUD12_HUMAN
|
Increases ADR
|
[6] |
|
Carbonic anhydrase-related protein 10 (CA10)
|
OTC3N1F6
|
CAH10_HUMAN
|
Increases ADR
|
[6] |
|
Actin-like protein 7B (ACTL7B)
|
OTCG3IGE
|
ACL7B_HUMAN
|
Increases ADR
|
[13] |
|
Growth/differentiation factor 3 (GDF3)
|
OTD3KGJK
|
GDF3_HUMAN
|
Increases ADR
|
[6] |
|
Microtubule-associated tumor suppressor candidate 2 (MTUS2)
|
OTDBDH2L
|
MTUS2_HUMAN
|
Increases ADR
|
[11] |
|
Coagulation factor V (F5)
|
OTDMZ3LT
|
FA5_HUMAN
|
Increases ADR
|
[14] |
|
Storkhead-box protein 2 (STOX2)
|
OTE9BK02
|
STOX2_HUMAN
|
Increases ADR
|
[6] |
|
Protein kinase C-binding protein NELL1 (NELL1)
|
OTF3TX3N
|
NELL1_HUMAN
|
Increases ADR
|
[6] |
|
Disabled homolog 2-interacting protein (DAB2IP)
|
OTF456VC
|
DAB2P_HUMAN
|
Increases ADR
|
[6] |
|
Dual specificity protein phosphatase CDC14C (CDC14C)
|
OTFJU7Y0
|
CC14C_HUMAN
|
Increases ADR
|
[6] |
|
Unconventional myosin-X (MYO10)
|
OTHB78ZQ
|
MYO10_HUMAN
|
Increases ADR
|
[6] |
|
Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PPARGC1A)
|
OTHCDQ22
|
PRGC1_HUMAN
|
Increases ADR
|
[6] |
|
Sodium- and chloride-dependent transporter XTRP3 (SLC6A20)
|
OTHRD4XO
|
S6A20_HUMAN
|
Increases ADR
|
[10] |
|
FYVE, RhoGEF and PH domain-containing protein 6 (FGD6)
|
OTI0T62C
|
FGD6_HUMAN
|
Increases ADR
|
[10] |
|
Insulin-like growth factor I (IGF1)
|
OTIIZR61
|
IGF1_HUMAN
|
Increases ADR
|
[6] |
|
Cytochrome P450 7B1 (CYP7B1)
|
OTIKFYP6
|
CP7B1_HUMAN
|
Increases ADR
|
[6] |
|
Pleckstrin homology-like domain family B member 1 (PHLDB1)
|
OTIRCB6I
|
PHLB1_HUMAN
|
Increases ADR
|
[6] |
|
Muscarinic acetylcholine receptor M3 (CHRM3)
|
OTKSD095
|
ACM3_HUMAN
|
Increases ADR
|
[6] |
|
Cytochrome P450 1A2 (CYP1A2)
|
OTLLBX48
|
CP1A2_HUMAN
|
Increases ADR
|
[15] |
|
Natural killer cells antigen CD94 (KLRD1)
|
OTMYLOV4
|
KLRD1_HUMAN
|
Increases ADR
|
[6] |
|
Retinoic acid receptor RXR-alpha (RXRA)
|
OTP1TBDM
|
RXRA_HUMAN
|
Increases ADR
|
[10] |
|
Kazrin (KAZN)
|
OTPM7BYM
|
KAZRN_HUMAN
|
Increases ADR
|
[10] |
|
Eyes absent homolog 2 (EYA2)
|
OTPVLA73
|
EYA2_HUMAN
|
Increases ADR
|
[6] |
|
Piezo-type mechanosensitive ion channel component 2 (PIEZO2)
|
OTQ7AT38
|
PIEZ2_HUMAN
|
Increases ADR
|
[6] |
|
PR domain zinc finger protein 1 (PRDM1)
|
OTQLSVBS
|
PRDM1_HUMAN
|
Increases ADR
|
[10] |
|
Glutamate receptor ionotropic, NMDA 3A (GRIN3A)
|
OTQS9GYY
|
NMD3A_HUMAN
|
Increases ADR
|
[6] |
|
Solute carrier family 35 member F1 (SLC35F1)
|
OTRXHR2P
|
S35F1_HUMAN
|
Increases ADR
|
[6] |
|
Glutamate receptor ionotropic, NMDA 2A (GRIN2A)
|
OTTP0KN8
|
NMDE1_HUMAN
|
Increases ADR
|
[6] |
|
Cytochrome c oxidase subunit 4 isoform 1, mitochondrial (COX4I1)
|
OTU0FC24
|
COX41_HUMAN
|
Increases ADR
|
[6] |
|
Oligodendrocyte transcription factor 3 (OLIG3)
|
OTU8XLAF
|
OLIG3_HUMAN
|
Increases ADR
|
[6] |
|
Protocadherin-15 (PCDH15)
|
OTU9C2EH
|
PCD15_HUMAN
|
Increases ADR
|
[6] |
|
Erlin-1 (ERLIN1)
|
OTUOOODY
|
ERLN1_HUMAN
|
Increases ADR
|
[6] |
|
Cancer/testis antigen 62 (CT62)
|
OTV9N4KM
|
CT62_HUMAN
|
Increases ADR
|
[6] |
|
Matrilin-2 (MATN2)
|
OTVDR68G
|
MATN2_HUMAN
|
Increases ADR
|
[10] |
|
PHD finger protein 10 (PHF10)
|
OTWDR4EX
|
PHF10_HUMAN
|
Increases ADR
|
[6] |
|
Ras GTPase-activating-like protein IQGAP2 (IQGAP2)
|
OTX2UA7P
|
IQGA2_HUMAN
|
Increases ADR
|
[6] |
|
NIPA-like protein 2 (NIPAL2)
|
OTX9UREP
|
NPAL2_HUMAN
|
Increases ADR
|
[6] |
|
EF-hand calcium-binding domain-containing protein 11 (EFCAB11)
|
OTXDJ9R8
|
EFC11_HUMAN
|
Increases ADR
|
[6] |
|
Juxtaposed with another zinc finger protein 1 (JAZF1)
|
OTXTYSYD
|
JAZF1_HUMAN
|
Increases ADR
|
[6] |
|
Adhesion G-protein coupled receptor G6 (ADGRG6)
|
OTY2UBXO
|
AGRG6_HUMAN
|
Increases ADR
|
[6] |
|
Branched-chain-amino-acid aminotransferase, cytosolic (BCAT1)
|
OTYHXLWZ
|
BCAT1_HUMAN
|
Increases ADR
|
[6] |
|
Aromatase (CYP19A1)
|
OTZ6XF74
|
CP19A_HUMAN
|
Increases ADR
|
[6] |
|
Myocyte-specific enhancer factor 2C (MEF2C)
|
OTZGF1Y5
|
MEF2C_HUMAN
|
Increases ADR
|
[6] |
|
Disintegrin and metalloproteinase domain-containing protein 12 (ADAM12)
|
OTZKOTDB
|
ADA12_HUMAN
|
Increases ADR
|
[6] |
|
F-box only protein 8 (FBXO8)
|
OTZNGJGW
|
FBX8_HUMAN
|
Increases ADR
|
[6] |
| ------------------------------------------------------------------------------------ |
|
|
|
|