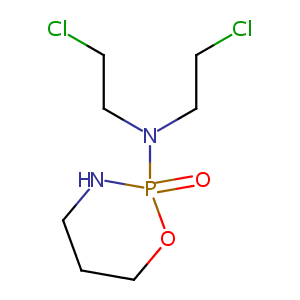
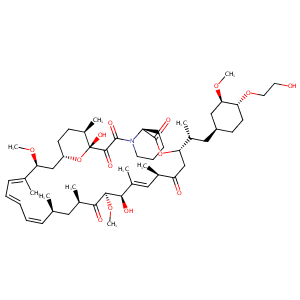
| DOT Name |
DOT ID |
UniProt ID |
Mode of Action |
REF |
|
Cytochrome P450 2C8 (CYP2C8)
|
OTHCWT42
|
CP2C8_HUMAN
|
Increases Activity
|
[23] |
|
Cytochrome P450 2C18 (CYP2C18)
|
OTY687L9
|
CP2CI_HUMAN
|
Increases Activity
|
[23] |
|
Cytochrome P450 3A4 (CYP3A4)
|
OTQGYY83
|
CP3A4_HUMAN
|
Increases Expression
|
[24] |
|
Cytochrome P450 2B6 (CYP2B6)
|
OTOYO4S7
|
CP2B6_HUMAN
|
Increases Hydroxylation
|
[25] |
|
Glucose-6-phosphate 1-dehydrogenase (G6PD)
|
OT300SMK
|
G6PD_HUMAN
|
Increases ADR
|
[8] |
|
Cytochrome P450 2D6 (CYP2D6)
|
OTZJC802
|
CP2D6_HUMAN
|
Increases Metabolism
|
[26] |
|
Cholinesterase (BCHE)
|
OTOH3WQ9
|
CHLE_HUMAN
|
Decreases Activity
|
[27] |
|
Actin, cytoplasmic 1 (ACTB)
|
OT1MCP2F
|
ACTB_HUMAN
|
Decreases Expression
|
[28] |
|
Cytochrome P450 1B1 (CYP1B1)
|
OTYXFLSD
|
CP1B1_HUMAN
|
Decreases Expression
|
[28] |
|
Band 4.1-like protein 2 (EPB41L2)
|
OT6UABPM
|
E41L2_HUMAN
|
Decreases Expression
|
[29] |
|
Receptor activity-modifying protein 1 (RAMP1)
|
OT7UT2XB
|
RAMP1_HUMAN
|
Increases Expression
|
[29] |
|
Cystatin-F (CST7)
|
OTQWZUVQ
|
CYTF_HUMAN
|
Decreases Expression
|
[29] |
|
Kelch repeat and BTB domain-containing protein 11 (KBTBD11)
|
OTBOY3WH
|
KBTBB_HUMAN
|
Decreases Expression
|
[29] |
|
Aldehyde dehydrogenase 1A1 (ALDH1A1)
|
OTCUWZKB
|
AL1A1_HUMAN
|
Increases Expression
|
[29] |
|
Carbonic anhydrase 2 (CA2)
|
OTJRMUAG
|
CAH2_HUMAN
|
Decreases Expression
|
[29] |
|
Aldehyde dehydrogenase, mitochondrial (ALDH2)
|
OTKJ9I3N
|
ALDH2_HUMAN
|
Increases Expression
|
[29] |
|
Tyrosine-protein kinase Fyn (FYN)
|
OTLSLVZS
|
FYN_HUMAN
|
Increases Expression
|
[29] |
|
Procathepsin L (CTSL)
|
OTYTUW29
|
CATL1_HUMAN
|
Increases Expression
|
[29] |
|
Galectin-1 (LGALS1)
|
OT8LDFWR
|
LEG1_HUMAN
|
Increases Expression
|
[29] |
|
SPARC (SPARC)
|
OTPN90H0
|
SPRC_HUMAN
|
Increases Expression
|
[29] |
|
Pro-cathepsin H (CTSH)
|
OTLFL0DG
|
CATH_HUMAN
|
Increases Expression
|
[29] |
|
Non-secretory ribonuclease (RNASE2)
|
OT8Z4FNE
|
RNAS2_HUMAN
|
Decreases Expression
|
[29] |
|
Transcriptional regulator ERG (ERG)
|
OTOTX9VU
|
ERG_HUMAN
|
Increases Expression
|
[29] |
|
Tissue factor (F3)
|
OT3MSU3B
|
TF_HUMAN
|
Decreases Expression
|
[29] |
|
Cytochrome c oxidase subunit 7A2, mitochondrial (COX7A2)
|
OTVT146E
|
CX7A2_HUMAN
|
Decreases Expression
|
[29] |
|
Methylated-DNA--protein-cysteine methyltransferase (MGMT)
|
OT40A9WH
|
MGMT_HUMAN
|
Decreases Expression
|
[29] |
|
Bactericidal permeability-increasing protein (BPI)
|
OTNN9LJ5
|
BPI_HUMAN
|
Decreases Expression
|
[29] |
|
Arachidonate 5-lipoxygenase-activating protein (ALOX5AP)
|
OT0DH40W
|
AL5AP_HUMAN
|
Decreases Expression
|
[29] |
|
Interferon-induced GTP-binding protein Mx2 (MX2)
|
OT05NF37
|
MX2_HUMAN
|
Decreases Expression
|
[29] |
|
Amine oxidase A (MAOA)
|
OT8NIWMQ
|
AOFA_HUMAN
|
Increases Expression
|
[29] |
|
Myeloblastin (PRTN3)
|
OT72MHP7
|
PRTN3_HUMAN
|
Decreases Expression
|
[29] |
|
Protein S100-P (S100P)
|
OTJCXNJG
|
S100P_HUMAN
|
Decreases Expression
|
[29] |
|
Caspase-1 (CASP1)
|
OTZ3YQFU
|
CASP1_HUMAN
|
Decreases Expression
|
[29] |
|
Myristoylated alanine-rich C-kinase substrate (MARCKS)
|
OT7N056G
|
MARCS_HUMAN
|
Increases Expression
|
[29] |
|
Transmembrane 4 L6 family member 1 (TM4SF1)
|
OTY0ECQN
|
T4S1_HUMAN
|
Increases Expression
|
[29] |
|
Intercellular adhesion molecule 3 (ICAM3)
|
OTTZ5A5D
|
ICAM3_HUMAN
|
Decreases Expression
|
[29] |
|
P2Y purinoceptor 2 (P2RY2)
|
OT47XN46
|
P2RY2_HUMAN
|
Decreases Expression
|
[29] |
|
Glycine receptor subunit beta (GLRB)
|
OTF37UG4
|
GLRB_HUMAN
|
Increases Expression
|
[29] |
|
C-C chemokine receptor type 8 (CCR8)
|
OTSCMH06
|
CCR8_HUMAN
|
Decreases Expression
|
[29] |
|
Ras-related protein R-Ras2 (RRAS2)
|
OT83NCEB
|
RRAS2_HUMAN
|
Increases Expression
|
[29] |
|
Bcl-2-interacting killer (BIK)
|
OTTH1T3D
|
BIK_HUMAN
|
Decreases Expression
|
[29] |
|
Angiopoietin-1 (ANGPT1)
|
OTVZ1NG3
|
ANGP1_HUMAN
|
Increases Expression
|
[29] |
|
Fascin (FSCN1)
|
OTTGIHTM
|
FSCN1_HUMAN
|
Increases Expression
|
[29] |
|
Pleckstrin homology-like domain family A member 2 (PHLDA2)
|
OTMV9DPP
|
PHLA2_HUMAN
|
Increases Expression
|
[29] |
|
Testis-expressed protein 30 (TEX30)
|
OT87WEPQ
|
TEX30_HUMAN
|
Decreases Expression
|
[29] |
|
Ras-related GTP-binding protein A (RRAGA)
|
OTKISLG4
|
RRAGA_HUMAN
|
Decreases Expression
|
[29] |
|
Cilium assembly protein DZIP1 (DZIP1)
|
OTBVPO66
|
DZIP1_HUMAN
|
Increases Expression
|
[29] |
|
RNA-binding protein with multiple splicing (RBPMS)
|
OT1RDKR9
|
RBPMS_HUMAN
|
Increases Expression
|
[29] |
|
Membrane-spanning 4-domains subfamily A member 3 (MS4A3)
|
OT0CEJOO
|
MS4A3_HUMAN
|
Decreases Expression
|
[29] |
|
Rho GTPase-activating protein 7 (DLC1)
|
OTP8LMCR
|
RHG07_HUMAN
|
Increases Expression
|
[29] |
|
MyoD family inhibitor domain-containing protein (MDFIC)
|
OTSRYBWZ
|
MDFIC_HUMAN
|
Decreases Expression
|
[29] |
|
Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-12 (GNG12)
|
OTOGF42G
|
GBG12_HUMAN
|
Increases Expression
|
[29] |
|
Cyclic AMP-dependent transcription factor ATF-5 (ATF5)
|
OT03QCLM
|
ATF5_HUMAN
|
Decreases Expression
|
[29] |
|
PALM2-AKAP2 fusion protein (PALM2AKAP2)
|
OTI618VF
|
PLAK2_HUMAN
|
Increases Expression
|
[29] |
|
Dihydropyrimidinase-related protein 4 (DPYSL4)
|
OT3SBS2S
|
DPYL4_HUMAN
|
Increases Expression
|
[9] |
|
Integral membrane protein GPR137B (GPR137B)
|
OT1CINQQ
|
G137B_HUMAN
|
Increases Expression
|
[9] |
|
Glutaminase kidney isoform, mitochondrial (GLS)
|
OTGOZG2M
|
GLSK_HUMAN
|
Increases Expression
|
[9] |
|
Urokinase-type plasminogen activator (PLAU)
|
OTX0QGKK
|
UROK_HUMAN
|
Increases Expression
|
[9] |
|
Ubiquitin-like protein ISG15 (ISG15)
|
OT53QQ7N
|
ISG15_HUMAN
|
Increases Expression
|
[9] |
|
Early growth response protein 1 (EGR1)
|
OTCP6XGZ
|
EGR1_HUMAN
|
Increases Expression
|
[9] |
|
Stomatin (STOM)
|
OTC8R6EH
|
STOM_HUMAN
|
Increases Expression
|
[9] |
|
Cellular retinoic acid-binding protein 2 (CRABP2)
|
OTY01V9G
|
RABP2_HUMAN
|
Increases Expression
|
[9] |
|
Tyrosine-protein kinase receptor UFO (AXL)
|
OTKA2SUX
|
UFO_HUMAN
|
Increases Expression
|
[9] |
|
14-3-3 protein sigma (SFN)
|
OTLJCZ1U
|
1433S_HUMAN
|
Increases Expression
|
[9] |
|
Cyclin-dependent kinase inhibitor 1 (CDKN1A)
|
OTQWHCZE
|
CDN1A_HUMAN
|
Increases Expression
|
[9] |
|
Tumor necrosis factor ligand superfamily member 9 (TNFSF9)
|
OTV9L89D
|
TNFL9_HUMAN
|
Increases Expression
|
[9] |
|
Galectin-7 (LGALS7)
|
OTMSVI7R
|
LEG7_HUMAN
|
Increases Expression
|
[9] |
|
Rho GDP-dissociation inhibitor 2 (ARHGDIB)
|
OT9PD6CS
|
GDIR2_HUMAN
|
Increases Expression
|
[9] |
|
Epithelial membrane protein 1 (EMP1)
|
OTSZHUHQ
|
EMP1_HUMAN
|
Increases Expression
|
[9] |
|
Epithelial membrane protein 3 (EMP3)
|
OTODMJ1D
|
EMP3_HUMAN
|
Increases Expression
|
[9] |
|
Protein ripply3 (RIPPLY3)
|
OT1HK35I
|
DSCR6_HUMAN
|
Increases Expression
|
[9] |
|
Pro-neuregulin-1, membrane-bound isoform (NRG1)
|
OTZO6F1X
|
NRG1_HUMAN
|
Increases Expression
|
[9] |
|
Antigen peptide transporter 1 (TAP1)
|
OTJL27PW
|
TAP1_HUMAN
|
Increases Expression
|
[9] |
|
Laminin subunit gamma-2 (LAMC2)
|
OTJMTM72
|
LAMC2_HUMAN
|
Increases Expression
|
[9] |
|
Filamin-C (FLNC)
|
OT3F8J6Y
|
FLNC_HUMAN
|
Increases Expression
|
[9] |
|
Immediate early response gene 5 protein (IER5)
|
OTJPTXMD
|
IER5_HUMAN
|
Increases Expression
|
[9] |
|
Keratin, type II cytoskeletal 80 (KRT80)
|
OTAU54U3
|
K2C80_HUMAN
|
Increases Expression
|
[9] |
|
Neuropilin and tolloid-like protein 2 (NETO2)
|
OT0YAMC0
|
NETO2_HUMAN
|
Increases Expression
|
[9] |
|
UDP-GalNAc:beta-1,3-N-acetylgalactosaminyltransferase 2 (B3GALNT2)
|
OTOF6O2B
|
B3GL2_HUMAN
|
Increases Expression
|
[9] |
|
Phospholipase ABHD3 (ABHD3)
|
OTH8P977
|
ABHD3_HUMAN
|
Increases Expression
|
[9] |
|
Tumor necrosis factor receptor superfamily member 14 (TNFRSF14)
|
OTB82PFO
|
TNR14_HUMAN
|
Increases Expression
|
[9] |
|
Major histocompatibility complex class I-related gene protein (MR1)
|
OTZU3XX7
|
HMR1_HUMAN
|
Increases Expression
|
[9] |
|
Tumor necrosis factor receptor superfamily member 27 (EDA2R)
|
OTJLAIIH
|
TNR27_HUMAN
|
Increases Expression
|
[9] |
|
Stathmin-3 (STMN3)
|
OT20F289
|
STMN3_HUMAN
|
Increases Expression
|
[9] |
|
Solute carrier family 2, facilitated glucose transporter member 6 (SLC2A6)
|
OTVUZCLG
|
GTR6_HUMAN
|
Increases Expression
|
[9] |
|
Endothelial protein C receptor (PROCR)
|
OTRHED17
|
EPCR_HUMAN
|
Increases Expression
|
[9] |
|
Serine/threonine-protein kinase Sgk1 (SGK1)
|
OT301T1U
|
SGK1_HUMAN
|
Decreases Expression
|
[10] |
|
Interferon-related developmental regulator 1 (IFRD1)
|
OT4SQMLQ
|
IFRD1_HUMAN
|
Decreases Expression
|
[10] |
|
Kinesin-like protein KIF21B (KIF21B)
|
OTFXFIU3
|
KI21B_HUMAN
|
Decreases Expression
|
[10] |
|
Large neutral amino acids transporter small subunit 3 (SLC43A1)
|
OTQJQY3S
|
LAT3_HUMAN
|
Decreases Expression
|
[10] |
|
Cystatin-A (CSTA)
|
OT1K68KE
|
CYTA_HUMAN
|
Decreases Expression
|
[10] |
|
Amino acid transporter heavy chain SLC3A2 (SLC3A2)
|
OTBR33M9
|
4F2_HUMAN
|
Decreases Expression
|
[10] |
|
Asparagine synthetase (ASNS)
|
OT8R922G
|
ASNS_HUMAN
|
Decreases Expression
|
[10] |
|
Endoplasmic reticulum chaperone BiP (HSPA5)
|
OTFUIRAO
|
BIP_HUMAN
|
Increases Expression
|
[10] |
|
Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial (MTHFD2)
|
OT1LQSGX
|
MTDC_HUMAN
|
Decreases Expression
|
[10] |
|
CCAAT/enhancer-binding protein beta (CEBPB)
|
OTM9MQIA
|
CEBPB_HUMAN
|
Decreases Expression
|
[10] |
|
Protein EVI2A (EVI2A)
|
OTR8RUXQ
|
EVI2A_HUMAN
|
Decreases Expression
|
[10] |
|
Tumor necrosis factor receptor superfamily member 6 (FAS)
|
OTP9XG86
|
TNR6_HUMAN
|
Decreases Expression
|
[10] |
|
DnaJ homolog subfamily B member 1 (DNAJB1)
|
OTCOSEVH
|
DNJB1_HUMAN
|
Increases Expression
|
[10] |
|
DNA damage-inducible transcript 3 protein (DDIT3)
|
OTI8YKKE
|
DDIT3_HUMAN
|
Decreases Expression
|
[10] |
|
B-cell lymphoma 6 protein (BCL6)
|
OTQAWWO1
|
BCL6_HUMAN
|
Decreases Expression
|
[10] |
|
Radiation-inducible immediate-early gene IEX-1 (IER3)
|
OTZJI5FZ
|
IEX1_HUMAN
|
Decreases Expression
|
[10] |
|
Cysteine--tRNA ligase, cytoplasmic (CARS1)
|
OTOUZF6O
|
SYCC_HUMAN
|
Decreases Expression
|
[10] |
|
Sestrin-2 (SESN2)
|
OT889IXY
|
SESN2_HUMAN
|
Decreases Expression
|
[10] |
|
Hydroxymethylglutaryl-CoA synthase, cytoplasmic (HMGCS1)
|
OTCO26FV
|
HMCS1_HUMAN
|
Increases Expression
|
[10] |
|
DNA-binding protein inhibitor ID-2 (ID2)
|
OT0U1D53
|
ID2_HUMAN
|
Decreases Expression
|
[10] |
|
Early activation antigen CD69 (CD69)
|
OTGJHSVP
|
CD69_HUMAN
|
Decreases Expression
|
[10] |
|
Transmembrane protein 267 (TMEM267)
|
OTEAAVK7
|
TM267_HUMAN
|
Decreases Expression
|
[10] |
|
Baculoviral IAP repeat-containing protein 3 (BIRC3)
|
OT3E95KB
|
BIRC3_HUMAN
|
Decreases Expression
|
[10] |
|
Isopentenyl-diphosphate Delta-isomerase 1 (IDI1)
|
OTGM06VJ
|
IDI1_HUMAN
|
Increases Expression
|
[10] |
|
N-arachidonyl glycine receptor (GPR18)
|
OTL2VFIV
|
GPR18_HUMAN
|
Decreases Expression
|
[10] |
|
Nuclear receptor coactivator 7 (NCOA7)
|
OT2CNBOG
|
NCOA7_HUMAN
|
Decreases Expression
|
[10] |
|
Alanine aminotransferase 2 (GPT2)
|
OTS5VF7N
|
ALAT2_HUMAN
|
Decreases Expression
|
[10] |
|
Nanos homolog 1 (NANOS1)
|
OT3UNZZY
|
NANO1_HUMAN
|
Increases Expression
|
[10] |
|
Heat shock protein 105 kDa (HSPH1)
|
OTVRR73T
|
HS105_HUMAN
|
Increases Expression
|
[10] |
|
Insulin gene enhancer protein ISL-2 (ISL2)
|
OT43PD2V
|
ISL2_HUMAN
|
Decreases Expression
|
[10] |
|
E3 ubiquitin-protein ligase pellino homolog 1 (PELI1)
|
OTMLBCLC
|
PELI1_HUMAN
|
Decreases Expression
|
[10] |
|
Centriolar and ciliogenesis-associated protein HYLS1 (HYLS1)
|
OT3SW5UC
|
HYLS1_HUMAN
|
Decreases Expression
|
[10] |
|
Tribbles homolog 3 (TRIB3)
|
OTG5OS7X
|
TRIB3_HUMAN
|
Decreases Expression
|
[10] |
|
Glutathione-specific gamma-glutamylcyclotransferase 1 (CHAC1)
|
OTJGE772
|
CHAC1_HUMAN
|
Decreases Expression
|
[10] |
|
Transmembrane 6 superfamily member 1 (TM6SF1)
|
OTQR4I2N
|
TM6S1_HUMAN
|
Decreases Expression
|
[10] |
|
Protein FAM107B (FAM107B)
|
OT5RG4J0
|
F107B_HUMAN
|
Decreases Expression
|
[10] |
|
Lysine-rich coiled-coil protein 1 (KRCC1)
|
OTMMV6WZ
|
KRCC1_HUMAN
|
Decreases Expression
|
[10] |
|
DnaJ homolog subfamily B member 4 (DNAJB4)
|
OTUD01BK
|
DNJB4_HUMAN
|
Increases Expression
|
[10] |
|
SLAM family member 5 (CD84)
|
OTAY5B0F
|
SLAF5_HUMAN
|
Increases Expression
|
[10] |
|
Cystine/glutamate transporter (SLC7A11)
|
OTKJ6PXW
|
XCT_HUMAN
|
Decreases Expression
|
[10] |
|
Nuclear factor erythroid 2-related factor 3 (NFE2L3)
|
OT1MGXT0
|
NF2L3_HUMAN
|
Decreases Expression
|
[10] |
|
Dual specificity protein phosphatase 10 (DUSP10)
|
OTNG467B
|
DUS10_HUMAN
|
Decreases Expression
|
[10] |
|
Apoptotic protease-activating factor 1 (APAF1)
|
OTJWIVY0
|
APAF_HUMAN
|
Decreases Expression
|
[11] |
|
Transmembrane protease serine 2 (TMPRSS2)
|
OTN44YQ5
|
TMPS2_HUMAN
|
Increases Expression
|
[30] |
|
Growth arrest-specific protein 2 (GAS2)
|
OT50JKXQ
|
GAS2_HUMAN
|
Increases Expression
|
[31] |
|
Lysine-specific histone demethylase 1A (KDM1A)
|
OT85JXS5
|
KDM1A_HUMAN
|
Increases Expression
|
[32] |
|
Slit homolog 1 protein (SLIT1)
|
OT35RBNT
|
SLIT1_HUMAN
|
Decreases Expression
|
[12] |
|
DnaJ homolog subfamily B member 6 (DNAJB6)
|
OTMHIIAN
|
DNJB6_HUMAN
|
Increases Expression
|
[31] |
|
Nuclear receptor subfamily 1 group I member 2 (NR1I2)
|
OTC5U0N5
|
NR1I2_HUMAN
|
Increases Activity
|
[33] |
|
Follitropin subunit beta (FSHB)
|
OTGLS283
|
FSHB_HUMAN
|
Increases Expression
|
[34] |
|
Thymidine kinase, cytosolic (TK1)
|
OTY5JFM1
|
KITH_HUMAN
|
Increases Mutagenesis
|
[35] |
|
Cellular tumor antigen p53 (TP53)
|
OTIE1VH3
|
P53_HUMAN
|
Increases Mutagenesis
|
[36] |
|
Endothelin-1 (EDN1)
|
OTZCACEG
|
EDN1_HUMAN
|
Increases Expression
|
[37] |
|
ATP-dependent translocase ABCB1 (ABCB1)
|
OTEJROBO
|
MDR1_HUMAN
|
Increases Expression
|
[33] |
|
Heme oxygenase 1 (HMOX1)
|
OTC1W6UX
|
HMOX1_HUMAN
|
Decreases Expression
|
[38] |
|
Poly polymerase 1 (PARP1)
|
OT310QSG
|
PARP1_HUMAN
|
Increases Cleavage
|
[39] |
|
Heat shock 70 kDa protein 1A (HSPA1A)
|
OTKGIE76
|
HS71A_HUMAN
|
Increases Expression
|
[40] |
|
Interleukin-8 (CXCL8)
|
OTS7T5VH
|
IL8_HUMAN
|
Increases Expression
|
[41] |
|
Apoptosis regulator Bcl-2 (BCL2)
|
OT9DVHC0
|
BCL2_HUMAN
|
Increases Expression
|
[42] |
|
Histone H2AX (H2AX)
|
OT18UX57
|
H2AX_HUMAN
|
Increases Phosphorylation
|
[43] |
|
Natriuretic peptides B (NPPB)
|
OTSN2IPY
|
ANFB_HUMAN
|
Increases Expression
|
[37] |
|
Troponin I, cardiac muscle (TNNI3)
|
OT65E12V
|
TNNI3_HUMAN
|
Increases Expression
|
[37] |
|
Tumor necrosis factor alpha-induced protein 3 (TNFAIP3)
|
OTVLI4DD
|
TNAP3_HUMAN
|
Increases Expression
|
[44] |
|
Growth arrest and DNA damage-inducible protein GADD45 alpha (GADD45A)
|
OTDRV63V
|
GA45A_HUMAN
|
Increases Expression
|
[11] |
|
Endothelin receptor type B (EDNRB)
|
OTLLZV3P
|
EDNRB_HUMAN
|
Increases Expression
|
[12] |
|
Insulin-like growth factor-binding protein 5 (IGFBP5)
|
OTRE5V0C
|
IBP5_HUMAN
|
Decreases Expression
|
[12] |
|
DNA cytosine-5)-methyltransferase 1 (DNMT1)
|
OTM2DGTK
|
DNMT1_HUMAN
|
Decreases Methylation
|
[32] |
|
Proteasome subunit alpha type-5 (PSMA5)
|
OT38E6Y1
|
PSA5_HUMAN
|
Increases Expression
|
[31] |
|
HLA class I histocompatibility antigen, alpha chain F (HLA-F)
|
OT76CM19
|
HLAF_HUMAN
|
Increases Expression
|
[12] |
|
Phospholipid hydroperoxide glutathione peroxidase GPX4 (GPX4)
|
OTRAFFX2
|
GPX4_HUMAN
|
Increases Expression
|
[31] |
|
Caspase-3 (CASP3)
|
OTIJRBE7
|
CASP3_HUMAN
|
Increases Expression
|
[42] |
|
Cytosolic purine 5'-nucleotidase (NT5C2)
|
OTJMF66Z
|
5NTC_HUMAN
|
Increases Expression
|
[12] |
|
Tumor protein D52 (TPD52)
|
OTPKSK43
|
TPD52_HUMAN
|
Decreases Expression
|
[44] |
|
Potassium voltage-gated channel subfamily KQT member 4 (KCNQ4)
|
OT29B58J
|
KCNQ4_HUMAN
|
Decreases Expression
|
[12] |
|
Heterogeneous nuclear ribonucleoprotein K (HNRNPK)
|
OTNPRM8U
|
HNRPK_HUMAN
|
Increases Expression
|
[31] |
|
Protein BTG2 (BTG2)
|
OTZF6K1H
|
BTG2_HUMAN
|
Increases Expression
|
[11] |
|
Runt-related transcription factor 3 (RUNX3)
|
OTITK1XD
|
RUNX3_HUMAN
|
Decreases Expression
|
[12] |
|
Calicin (CCIN)
|
OTRFZOT0
|
CALI_HUMAN
|
Increases Expression
|
[31] |
|
Programmed cell death protein 4 (PDCD4)
|
OTZ6NXUX
|
PDCD4_HUMAN
|
Decreases Expression
|
[44] |
|
Ribonucleoside-diphosphate reductase subunit M2 B (RRM2B)
|
OTE8GBUR
|
RIR2B_HUMAN
|
Increases Expression
|
[45] |
|
Partitioning defective 3 homolog (PARD3)
|
OTH5BPLO
|
PARD3_HUMAN
|
Decreases Expression
|
[12] |
|
GATOR1 complex protein NPRL2 (NPRL2)
|
OTOB10MO
|
NPRL2_HUMAN
|
Decreases Expression
|
[12] |
|
F-box/WD repeat-containing protein 7 (FBXW7)
|
OTJXE4OT
|
FBXW7_HUMAN
|
Increases Expression
|
[11] |
|
Rho guanine nucleotide exchange factor 26 (ARHGEF26)
|
OTX0U8PX
|
ARHGQ_HUMAN
|
Increases Expression
|
[12] |
|
G-protein coupled receptor 20 (GPR20)
|
OTPCR8F7
|
GPR20_HUMAN
|
Increases Expression
|
[12] |
|
Angiotensin-converting enzyme 2 (ACE2)
|
OTTRZGU7
|
ACE2_HUMAN
|
Decreases Expression
|
[30] |
|
Polyadenylate-binding protein-interacting protein 1 (PAIP1)
|
OTL2F5T5
|
PAIP1_HUMAN
|
Decreases Expression
|
[12] |
|
Large subunit GTPase 1 homolog (LSG1)
|
OTDAXJ9L
|
LSG1_HUMAN
|
Decreases Expression
|
[12] |
|
Glyoxalase domain-containing protein 4 (GLOD4)
|
OTBJKFXA
|
GLOD4_HUMAN
|
Decreases Expression
|
[12] |
|
DNA-directed DNA/RNA polymerase mu (POLM)
|
OT0SRIP4
|
DPOLM_HUMAN
|
Increases Expression
|
[12] |
|
Ubiquitin-like-conjugating enzyme ATG3 (ATG3)
|
OT28VBVK
|
ATG3_HUMAN
|
Decreases Expression
|
[12] |
|
Regulator of telomere elongation helicase 1 (RTEL1)
|
OTI3PJCT
|
RTEL1_HUMAN
|
Increases Expression
|
[31] |
|
FK506-binding protein-like (FKBPL)
|
OTR9ND6K
|
FKBPL_HUMAN
|
Increases Expression
|
[42] |
|
Mitochondrial pyruvate carrier 2 (MPC2)
|
OT0GHXGG
|
MPC2_HUMAN
|
Affects Response To Substance
|
[13] |
|
Krueppel-like factor 5 (KLF5)
|
OT1ABI9N
|
KLF5_HUMAN
|
Affects Response To Substance
|
[13] |
|
Galactocerebrosidase (GALC)
|
OT1F6BZK
|
GALC_HUMAN
|
Affects Response To Substance
|
[13] |
|
Granulocyte-macrophage colony-stimulating factor (CSF2)
|
OT1M7D28
|
CSF2_HUMAN
|
Affects Activity
|
[46] |
|
Superoxide dismutase (SOD1)
|
OT39TA1L
|
SODC_HUMAN
|
Increases ADR
|
[8] |
|
Krueppel-like factor 4 (KLF4)
|
OT4O9RQW
|
KLF4_HUMAN
|
Decreases Response To Substance
|
[47] |
|
D-beta-hydroxybutyrate dehydrogenase, mitochondrial (BDH1)
|
OT62RL5P
|
BDH_HUMAN
|
Affects Response To Substance
|
[13] |
|
Melanoma-associated antigen D1 (MAGED1)
|
OT6EOLFC
|
MAGD1_HUMAN
|
Affects Response To Substance
|
[13] |
|
Prostaglandin G/H synthase 2 (PTGS2)
|
OT75U9M4
|
PGH2_HUMAN
|
Affects Response To Substance
|
[13] |
|
Pro-adrenomedullin (ADM)
|
OT7T0TA4
|
ADML_HUMAN
|
Affects Response To Substance
|
[13] |
|
Peptidyl-prolyl cis-trans isomerase FKBP1B (FKBP1B)
|
OT8CMPB2
|
FKB1B_HUMAN
|
Affects Response To Substance
|
[13] |
|
L-lactate dehydrogenase B chain (LDHB)
|
OT9B1CT3
|
LDHB_HUMAN
|
Affects Response To Substance
|
[13] |
|
Epsilon-sarcoglycan (SGCE)
|
OT9F17JB
|
SGCE_HUMAN
|
Affects Response To Substance
|
[13] |
|
Caspase-8 (CASP8)
|
OTA8TVI8
|
CASP8_HUMAN
|
Increases Response To Substance
|
[48] |
|
Aldehyde dehydrogenase, dimeric NADP-preferring (ALDH3A1)
|
OTAYZZE6
|
AL3A1_HUMAN
|
Affects Response To Substance
|
[49] |
|
Sterol O-acyltransferase 1 (SOAT1)
|
OTB4Y5RJ
|
SOAT1_HUMAN
|
Affects Response To Substance
|
[13] |
|
Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3)
|
OTB97VIK
|
IF2B3_HUMAN
|
Affects Response To Substance
|
[13] |
|
Cochlin (COCH)
|
OTBEHD89
|
COCH_HUMAN
|
Affects Response To Substance
|
[13] |
|
Fibronectin type III domain-containing protein 3B (FNDC3B)
|
OTBILGDR
|
FND3B_HUMAN
|
Affects Response To Substance
|
[13] |
|
Tropomyosin alpha-1 chain (TPM1)
|
OTD73X6R
|
TPM1_HUMAN
|
Affects Response To Substance
|
[13] |
|
Very long chain fatty acid elongase 2 (ELOVL2)
|
OTDAF6U3
|
ELOV2_HUMAN
|
Affects Response To Substance
|
[13] |
|
Glutathione peroxidase 1 (GPX1)
|
OTE2O72Q
|
GPX1_HUMAN
|
Increases ADR
|
[8] |
|
Protein AF1q (MLLT11)
|
OTG5RVHC
|
AF1Q_HUMAN
|
Decreases Response To Substance
|
[50] |
|
Troponin I, fast skeletal muscle (TNNI2)
|
OTGGZFSC
|
TNNI2_HUMAN
|
Increases ADR
|
[8] |
|
Multidrug resistance-associated protein 1 (ABCC1)
|
OTGUN89S
|
MRP1_HUMAN
|
Decreases Response To Substance
|
[51] |
|
Tomoregulin-1 (TMEFF1)
|
OTH6M3CH
|
TEFF1_HUMAN
|
Affects Response To Substance
|
[13] |
|
DNA damage-inducible transcript 4 protein (DDIT4)
|
OTHY8SY4
|
DDIT4_HUMAN
|
Affects Response To Substance
|
[13] |
|
Ribosomal protein S6 kinase alpha-2 (RPS6KA2)
|
OTIOYUSU
|
KS6A2_HUMAN
|
Affects Response To Substance
|
[13] |
|
Superoxide dismutase , mitochondrial (SOD2)
|
OTIWXGZ9
|
SODM_HUMAN
|
Affects Response To Substance
|
[52] |
|
Insulin-like growth factor-binding protein 3 (IGFBP3)
|
OTIX63TX
|
IBP3_HUMAN
|
Affects Response To Substance
|
[13] |
|
Osteopontin (SPP1)
|
OTJGC23Y
|
OSTP_HUMAN
|
Affects Response To Substance
|
[13] |
|
Heat shock cognate 71 kDa protein (HSPA8)
|
OTJI2RCI
|
HSP7C_HUMAN
|
Affects Response To Substance
|
[13] |
|
Nidogen-1 (NID1)
|
OTKLBLS6
|
NID1_HUMAN
|
Affects Response To Substance
|
[13] |
|
Glutathione reductase, mitochondrial (GSR)
|
OTM2TUYM
|
GSHR_HUMAN
|
Increases ADR
|
[8] |
|
Homeobox protein Hox-A1 (HOXA1)
|
OTMSOJ7D
|
HXA1_HUMAN
|
Affects Response To Substance
|
[13] |
|
Trypsin-3 (PRSS3)
|
OTN3S5YB
|
TRY3_HUMAN
|
Affects Response To Substance
|
[13] |
|
Melanoma-associated antigen D4 (MAGED4B)
|
OTO37U7W
|
MAGD4_HUMAN
|
Affects Response To Substance
|
[13] |
|
Opsin-3 (OPN3)
|
OTON6BFU
|
OPN3_HUMAN
|
Affects Response To Substance
|
[13] |
|
Interleukin-1 alpha (IL1A)
|
OTPSGILV
|
IL1A_HUMAN
|
Affects Response To Substance
|
[53] |
|
Complement factor I (CFI)
|
OTQYYX0P
|
CFAI_HUMAN
|
Affects Response To Substance
|
[13] |
|
Heat shock-related 70 kDa protein 2 (HSPA2)
|
OTSDET7B
|
HSP72_HUMAN
|
Affects Response To Substance
|
[13] |
|
Cytochrome P450 3A5 (CYP3A5)
|
OTSXFBXB
|
CP3A5_HUMAN
|
Affects Response To Substance
|
[54] |
|
Max-interacting protein 1 (MXI1)
|
OTUQ9E0D
|
MXI1_HUMAN
|
Affects Response To Substance
|
[13] |
|
Glycogen debranching enzyme (AGL)
|
OTWBM7WY
|
GDE_HUMAN
|
Affects Response To Substance
|
[13] |
|
Myosin-10 (MYH10)
|
OTXN2WXS
|
MYH10_HUMAN
|
Affects Response To Substance
|
[13] |
|
Sal-like protein 1 (SALL1)
|
OTYYZGLH
|
SALL1_HUMAN
|
Affects Response To Substance
|
[13] |
| ------------------------------------------------------------------------------------ |
|
|
|
|