| 1 |
Loss of function mutations in VARS encoding cytoplasmic valyl-tRNA synthetase cause microcephaly, seizures, and progressive cerebral atrophy.Hum Genet. 2018 Apr;137(4):293-303. doi: 10.1007/s00439-018-1882-3. Epub 2018 Apr 24.
|
| 2 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5692).
|
| 3 |
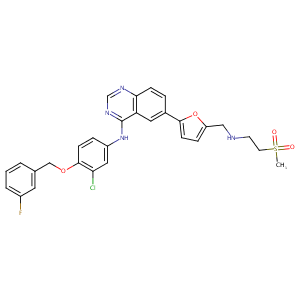
Lapatinib FDA Label
|
| 4 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7143).
|
| 5 |
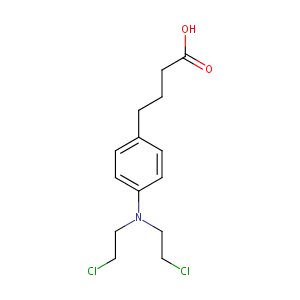
Chlorambucil FDA Label
|
| 6 |
UGT-dependent regioselective glucuronidation of ursodeoxycholic acid and obeticholic acid and selective transport of the consequent acyl glucuronides by OATP1B1 and 1B3. Chem Biol Interact. 2019 Sep 1;310:108745. doi: 10.1016/j.cbi.2019.108745. Epub 2019 Jul 9.
|
| 7 |
Triple negative breast cancer--current status and prospective targeted treatment based on HER1 (EGFR), TOP2A and C-MYC gene assessment. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2009 Mar;153(1):13-7.
|
| 8 |
Inhibition of eEF-2 kinase sensitizes human nasopharyngeal carcinoma cells to lapatinib-induced apoptosis through the Src and Erk pathways.BMC Cancer. 2016 Oct 19;16(1):813.
|
| 9 |
Tarascon Pocket Pharmacopoeia 2018 Classic Shirt-Pocket Edition.
|
| 10 |
The role of efflux and uptake transporters in [N-{3-chloro-4-[(3-fluorobenzyl)oxy]phenyl}-6-[5-({[2-(methylsulfonyl)ethyl]amino}methyl)-2-furyl]-4-quinazolinamine (GW572016, lapatinib) disposition and drug interactions. Drug Metab Dispos. 2008 Apr;36(4):695-701.
|
| 11 |
Mechanism-based inactivation of cytochrome P450 3A4 by lapatinib. Mol Pharmacol. 2010 Oct;78(4):693-703.
|
| 12 |
Clinical pharmacokinetics of tyrosine kinase inhibitors. Cancer Treat Rev. 2009 Dec;35(8):692-706.
|
| 13 |
The dual ErbB1/ErbB2 inhibitor, lapatinib (GW572016), cooperates with tamoxifen to inhibit both cell proliferation- and estrogen-dependent gene expression in antiestrogen-resistant breast cancer. Cancer Res. 2005 Jan 1;65(1):18-25.
|
| 14 |
Interference with bile salt export pump function is a susceptibility factor for human liver injury in drug development. Toxicol Sci. 2010 Dec; 118(2):485-500.
|
| 15 |
P450 3A-catalyzed O-dealkylation of lapatinib induces mitochondrial stress and activates Nrf2. Chem Res Toxicol. 2016 May 16;29(5):784-96.
|
| 16 |
Combining lapatinib (GW572016), a small molecule inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing breast cancer cells. Oncogene. 2005 Sep 15;24(41):6213-21. doi: 10.1038/sj.onc.1208774.
|
| 17 |
CDK4/6 inhibition provides a potent adjunct to Her2-targeted therapies in preclinical breast cancer models. Genes Cancer. 2014 Jul;5(7-8):261-72. doi: 10.18632/genesandcancer.24.
|
| 18 |
Effects of lapatinib on cell proliferation and apoptosis in NB4 cells. Oncol Lett. 2018 Jan;15(1):235-242. doi: 10.3892/ol.2017.7342. Epub 2017 Nov 3.
|
| 19 |
The involvement of hepatic cytochrome P450s in the cytotoxicity of lapatinib. Toxicol Sci. 2023 Dec 21;197(1):69-78. doi: 10.1093/toxsci/kfad099.
|
| 20 |
Evaluating the Role of Multidrug Resistance Protein 3 (MDR3) Inhibition in Predicting Drug-Induced Liver Injury Using 125 Pharmaceuticals. Chem Res Toxicol. 2017 May 15;30(5):1219-1229. doi: 10.1021/acs.chemrestox.7b00048. Epub 2017 May 4.
|
| 21 |
Suppression of HER2/HER3-mediated growth of breast cancer cells with combinations of GDC-0941 PI3K inhibitor, trastuzumab, and pertuzumab. Clin Cancer Res. 2009 Jun 15;15(12):4147-56. doi: 10.1158/1078-0432.CCR-08-2814. Epub 2009 Jun 9.
|
| 22 |
Cytotoxicity of 34 FDA approved small-molecule kinase inhibitors in primary rat and human hepatocytes. Toxicol Lett. 2018 Jul;291:138-148. doi: 10.1016/j.toxlet.2018.04.010. Epub 2018 Apr 12.
|
| 23 |
Association of CYP1A1 and CYP1B1 inhibition in in vitro assays with drug-induced liver injury. J Toxicol Sci. 2021;46(4):167-176. doi: 10.2131/jts.46.167.
|
| 24 |
The K-Ras effector p38 MAPK confers intrinsic resistance to tyrosine kinase inhibitors by stimulating EGFR transcription and EGFR dephosphorylation. J Biol Chem. 2017 Sep 8;292(36):15070-15079. doi: 10.1074/jbc.M117.779488. Epub 2017 Jul 24.
|
| 25 |
HLA-DQA1*02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J Clin Oncol. 2011 Feb 20;29(6):667-73. doi: 10.1200/JCO.2010.31.3197. Epub 2011 Jan 18.
|
| 26 |
Niclosamide inhibits epithelial-mesenchymal transition and tumor growth in lapatinib-resistant human epidermal growth factor receptor 2-positive breast cancer. Int J Biochem Cell Biol. 2016 Feb;71:12-23. doi: 10.1016/j.biocel.2015.11.014. Epub 2015 Nov 28.
|
| 27 |
Roles of DNA repair and reductase activity in the cytotoxicity of the hypoxia-activated dinitrobenzamide mustard PR-104A. Mol Cancer Ther. 2009 Jun;8(6):1714-23.
|
| 28 |
Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016 Jan 1;370(1):153-64.
|
| 29 |
Transporters and renal drug elimination. Annu Rev Pharmacol Toxicol. 2004;44:137-66.
|
| 30 |
The anti-cancer drug chlorambucil as a substrate for the human polymorphic enzyme glutathione transferase P1-1: kinetic properties and crystallographic characterisation of allelic variants. J Mol Biol. 2008 Jun 27;380(1):131-44.
|
| 31 |
The influence of coordinate overexpression of glutathione phase II detoxification gene products on drug resistance. J Pharmacol Exp Ther. 2000 Aug;294(2):480-7.
|
| 32 |
Enhanced in vitro invasiveness and drug resistance with altered gene expression patterns in a human lung carcinoma cell line after pulse selection with anticancer drugs. Int J Cancer. 2004 Sep 10;111(4):484-93. doi: 10.1002/ijc.20230.
|
| 33 |
Role of multidrug resistance protein 1 (MRP1) and glutathione S-transferase A1-1 in alkylating agent resistanceKinetics of glutathione conjugate formation and efflux govern differential cellular sensitivity to chlorambucil versus melphalan toxicity. J Biol Chem. 2001 Mar 16;276(11):7952-6.
|
| 34 |
Oxidative stress mechanisms do not discriminate between genotoxic and nongenotoxic liver carcinogens. Chem Res Toxicol. 2015 Aug 17;28(8):1636-46.
|
| 35 |
Role of the TRAIL/APO2-L death receptors in chlorambucil- and fludarabine-induced apoptosis in chronic lymphocytic leukemia. Oncogene. 2003 Nov 13;22(51):8356-69. doi: 10.1038/sj.onc.1207004.
|
| 36 |
Differential effects of chemotherapeutic drugs versus the MDM-2 antagonist nutlin-3 on cell cycle progression and induction of apoptosis in SKW6.4 lymphoblastoid B-cells. J Cell Biochem. 2008 May 15;104(2):595-605. doi: 10.1002/jcb.21649.
|
| 37 |
Differential gene expression induction by TRAIL in B chronic lymphocytic leukemia (B-CLL) cells showing high versus low levels of Zap-70. J Cell Physiol. 2007 Oct;213(1):229-36. doi: 10.1002/jcp.21116.
|
| 38 |
Theophylline synergizes with chlorambucil in inducing apoptosis of B-chronic lymphocytic leukemia cells. Blood. 1996 Sep 15;88(6):2172-82.
|
| 39 |
A two-hit mechanism for pre-mitotic arrest of cancer cell proliferation by a polyamide-alkylator conjugate. Cell Cycle. 2006 Jul;5(14):1537-48. doi: 10.4161/cc.5.14.2913. Epub 2006 Jul 17.
|
| 40 |
Systems pharmacological analysis of drugs inducing stevens-johnson syndrome and toxic epidermal necrolysis. Chem Res Toxicol. 2015 May 18;28(5):927-34. doi: 10.1021/tx5005248. Epub 2015 Apr 3.
|
| 41 |
Caspase 8 activation independent of Fas (CD95/APO-1) signaling may mediate killing of B-chronic lymphocytic leukemia cells by cytotoxic drugs or gamma radiation. Blood. 2001 Nov 1;98(9):2800-7. doi: 10.1182/blood.v98.9.2800.
|
| 42 |
Disruption of gene expression and induction of apoptosis in prostate cancer cells by a DNA-damaging agent tethered to an androgen receptor ligand. Chem Biol. 2005 Jul;12(7):779-87. doi: 10.1016/j.chembiol.2005.05.009.
|
| 43 |
Chlorambucil induction of HsRad51 in B-cell chronic lymphocytic leukemia. Clin Cancer Res. 1999 Aug;5(8):2178-84.
|
| 44 |
Bcl-2 antisense oligonucleotides enhance the cytotoxicity of chlorambucil in B-cell chronic lymphocytic leukaemia cells. Leuk Lymphoma. 2001 Jul;42(3):491-8. doi: 10.3109/10428190109064606.
|
| 45 |
The MT1G Gene in LUHMES Neurons Is a Sensitive Biomarker of Neurotoxicity. Neurotox Res. 2020 Dec;38(4):967-978. doi: 10.1007/s12640-020-00272-3. Epub 2020 Sep 1.
|
| 46 |
Characterization of the cancer chemopreventive NRF2-dependent gene battery in human keratinocytes: demonstration that the KEAP1-NRF2 pathway, and not the BACH1-NRF2 pathway, controls cytoprotection against electrophiles as well as redox-cycling compounds. Carcinogenesis. 2009 Sep;30(9):1571-80. doi: 10.1093/carcin/bgp176. Epub 2009 Jul 16.
|
| 47 |
Increased resistance to nitrogen mustards and antifolates following in vitro selection of murine fibroblasts and primary hematopoietic cells transduced with a bicistronic retroviral vector expressing the rat glutathione S-transferase A3 and a mutant dihydrofolate reductase. Cancer Gene Ther. 2003 Aug;10(8):637-46. doi: 10.1038/sj.cgt.7700619.
|
| 48 |
Coexpression of rat glutathione S-transferase A3 and human cytidine deaminase by a bicistronic retroviral vector confers in vitro resistance to nitrogen mustards and cytosine arabinoside in murine fibroblasts. Cancer Gene Ther. 2000 May;7(5):757-65. doi: 10.1038/sj.cgt.7700169.
|
| 49 |
In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005 Oct 15;11(20):7508-15. doi: 10.1158/1078-0432.CCR-05-1048.
|
| 50 |
Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res. 2000 May;6(5):1796-803.
|
| 51 |
Role of multidrug resistance protein 2 (MRP2, ABCC2) in alkylating agent detoxification: MRP2 potentiates glutathione S-transferase A1-1-mediated resistance to chlorambucil cytotoxicity. J Pharmacol Exp Ther. 2004 Jan;308(1):260-7. doi: 10.1124/jpet.103.057729. Epub 2003 Oct 20.
|
| 52 |
Glutathione S-transferase M1 and multidrug resistance protein 1 act in synergy to protect melanoma cells from vincristine effects. Mol Pharmacol. 2004 Apr;65(4):897-905. doi: 10.1124/mol.65.4.897.
|
|
|
|
|
|
|