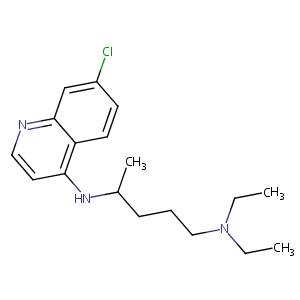
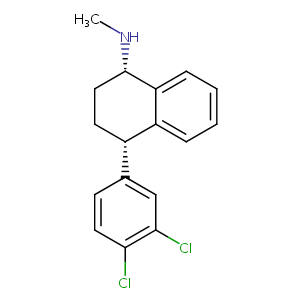
| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5535).
|
| 3 |
Chloroquine FDA Label
|
| 4 |
ClinicalTrials.gov (NCT04360759) Chloroquine Outpatient Treatment Evaluation for HIV-Covid-19. U.S. National Institutes of Health.
|
| 5 |
Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016 May;15(5):327-47.
|
| 6 |
Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov. 2020 Mar;19(3):149-150.
|
| 7 |
Sertraline FDA Label
|
| 8 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 4798).
|
| 9 |
Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020 Mar;30(3):269-271.
|
| 10 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services.
|
| 11 |
Improving the prediction of the brain disposition for orally administered drugs using BDDCS. Adv Drug Deliv Rev. 2012 Jan;64(1):95-109.
|
| 12 |
Molecular mechanism of renal tubular secretion of the antimalarial drug chloroquine. Antimicrob Agents Chemother. 2011 Jul;55(7):3091-8.
|
| 13 |
Cytochrome P450 2C8 and CYP3A4/5 are involved in chloroquine metabolism in human liver microsomes. Arch Pharm Res. 2003 Aug;26(8):631-7.
|
| 14 |
In vitro metabolism of chloroquine: identification of CYP2C8, CYP3A4, and CYP2D6 as the main isoforms catalyzing N-desethylchloroquine formation. Drug Metab Dispos. 2003 Jun;31(6):748-54.
|
| 15 |
Halofantrine and chloroquine inhibit CYP2D6 activity in healthy Zambians. Br J Clin Pharmacol. 1998 Mar;45(3):315-7.
|
| 16 |
Short communication: high prevalence of the cytochrome P450 2C8*2 mutation in Northern Ghana. Trop Med Int Health. 2005 Dec;10(12):1271-3.
|
| 17 |
Application of higher throughput screening (HTS) inhibition assays to evaluate the interaction of antiparasitic drugs with cytochrome P450s. Drug Metab Dispos. 2001 Jan;29(1):30-5.
|
| 18 |
Autophagy inhibition upregulates CD4(+) tumor infiltrating lymphocyte expression via miR-155 regulation and TRAIL activation. Mol Oncol. 2016 Dec;10(10):1516-1531. doi: 10.1016/j.molonc.2016.08.005. Epub 2016 Sep 16.
|
| 19 |
Niclosamide, an anti-helminthic molecule, downregulates the retroviral oncoprotein Tax and pro-survival Bcl-2 proteins in HTLV-1-transformed T lymphocytes. Biochem Biophys Res Commun. 2015 Aug 14;464(1):221-228. doi: 10.1016/j.bbrc.2015.06.120. Epub 2015 Jun 23.
|
| 20 |
Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435-1455. doi: 10.1080/15548627.2018.1474314. Epub 2018 Jul 20.
|
| 21 |
Chloroquine and Hydroxychloroquine Increase Retinal Pigment Epithelial Layer Permeability. J Biochem Mol Toxicol. 2015 Jul;29(7):299-304. doi: 10.1002/jbt.21696. Epub 2015 Mar 9.
|
| 22 |
Sulindac metabolites induce proteosomal and lysosomal degradation of the epidermal growth factor receptor. Cancer Prev Res (Phila). 2010 Apr;3(4):560-72. doi: 10.1158/1940-6207.CAPR-09-0159. Epub 2010 Mar 23.
|
| 23 |
Artesunate alleviates liver fibrosis by regulating ferroptosis signaling pathway. Biomed Pharmacother. 2019 Jan;109:2043-2053. doi: 10.1016/j.biopha.2018.11.030. Epub 2018 Nov 26.
|
| 24 |
Calcitriol-mediated hypercalcaemia and increased interleukins in a patient with sarcoid myopathy. Clin Rheumatol. 1999;18(6):488-91. doi: 10.1007/s100670050144.
|
| 25 |
Reactive oxygen species mediate chloroquine-induced expression of chemokines by human astroglial cells. Glia. 2004 Jul;47(1):9-20. doi: 10.1002/glia.20017.
|
| 26 |
Profiling the immunotoxicity of chemicals based on in vitro evaluation by a combination of the Multi-ImmunoTox assay and the IL-8 Luc assay. Arch Toxicol. 2018 Jun;92(6):2043-2054. doi: 10.1007/s00204-018-2199-7. Epub 2018 Mar 29.
|
| 27 |
High-throughput measurement of the Tp53 response to anticancer drugs and random compounds using a stably integrated Tp53-responsive luciferase reporter. Carcinogenesis. 2002 Jun;23(6):949-57. doi: 10.1093/carcin/23.6.949.
|
| 28 |
Downregulation of Organic Anion Transporting Polypeptide (OATP) 1B1 Transport Function by Lysosomotropic Drug Chloroquine: Implication in OATP-Mediated Drug-Drug Interactions. Mol Pharm. 2016 Mar 7;13(3):839-51. doi: 10.1021/acs.molpharmaceut.5b00763. Epub 2016 Feb 1.
|
| 29 |
Protecting cells by protecting their vulnerable lysosomes: Identification of a new mechanism for preserving lysosomal functional integrity upon oxidative stress. PLoS Genet. 2017 Feb 9;13(2):e1006603. doi: 10.1371/journal.pgen.1006603. eCollection 2017 Feb.
|
| 30 |
Chloroquine aggravates the arsenic trioxide (As2O3)-induced apoptosis of acute promyelocytic leukemia NB4 cells via inhibiting lysosomal degradation in vitro. Eur Rev Med Pharmacol Sci. 2018 Oct;22(19):6412-6421. doi: 10.26355/eurrev_201810_16054.
|
| 31 |
Chloroquine has tumor-inhibitory and tumor-promoting effects in triple-negative breast cancer. Oncol Lett. 2013 Dec;6(6):1665-1672. doi: 10.3892/ol.2013.1602. Epub 2013 Oct 4.
|
| 32 |
NDRG1 inhibition sensitizes osteosarcoma cells to combretastatin A-4 through targeting autophagy. Cell Death Dis. 2017 Sep 14;8(9):e3048. doi: 10.1038/cddis.2017.438.
|
| 33 |
Toxicoproteomics reveals an effect of clozapine on autophagy in human liver spheroids. Toxicol Mech Methods. 2023 Jun;33(5):401-410. doi: 10.1080/15376516.2022.2156005. Epub 2022 Dec 19.
|
| 34 |
Increasing intratumor C/EBP- LIP and nitric oxide levels overcome resistance to doxorubicin in triple negative breast cancer. J Exp Clin Cancer Res. 2018 Nov 27;37(1):286. doi: 10.1186/s13046-018-0967-0.
|
| 35 |
Inhibition of autophagic flux differently modulates cannabidiol-induced death in 2D and 3D glioblastoma cell cultures. Sci Rep. 2020 Feb 14;10(1):2687. doi: 10.1038/s41598-020-59468-4.
|
| 36 |
Effect of toll-like receptor 7 and 9 targeted therapy to prevent the development of hepatocellular carcinoma. Liver Int. 2015 Mar;35(3):1063-76. doi: 10.1111/liv.12626. Epub 2014 Jul 30.
|
| 37 |
Control of mammary tumor cell growth in vitro by novel cell differentiation and apoptosis agents. Breast Cancer Res Treat. 2002 Sep;75(2):107-17. doi: 10.1023/a:1019698807564.
|
| 38 |
Chronic use of chloroquine disrupts the urine concentration mechanism by lowering cAMP levels in the inner medulla. Am J Physiol Renal Physiol. 2012 Sep 15;303(6):F900-5. doi: 10.1152/ajprenal.00547.2011. Epub 2012 Jul 11.
|
| 39 |
The dual PI3K/mTOR inhibitor NVP-BEZ235 and chloroquine synergize to trigger apoptosis via mitochondrial-lysosomal cross-talk. Int J Cancer. 2013 Jun 1;132(11):2682-93. doi: 10.1002/ijc.27935. Epub 2012 Dec 4.
|
| 40 |
Paradoxical Effect of Chloroquine Treatment in Enhancing Chikungunya Virus Infection. Viruses. 2018 May 17;10(5):268. doi: 10.3390/v10050268.
|
| 41 |
Cadmium induces triglyceride levels via microsomal triglyceride transfer protein (MTTP) accumulation caused by lysosomal deacidification regulated by endoplasmic reticulum (ER) Ca(2+) homeostasis. Chem Biol Interact. 2021 Oct 1;348:109649. doi: 10.1016/j.cbi.2021.109649. Epub 2021 Sep 10.
|
| 42 |
Chloroquine inhibits T cell proliferation by interfering with IL-2 production and responsiveness. Clin Exp Immunol. 1995 Oct;102(1):144-51. doi: 10.1111/j.1365-2249.1995.tb06648.x.
|
| 43 |
Presenilin-2 regulates the degradation of RBP-Jk protein through p38 mitogen-activated protein kinase. J Cell Sci. 2012 Mar 1;125(Pt 5):1296-308. doi: 10.1242/jcs.095984. Epub 2012 Feb 2.
|
| 44 |
Inhibition of cholesterol metabolism underlies synergy between mTOR pathway inhibition and chloroquine in bladder cancer cells. Oncogene. 2016 Aug 25;35(34):4518-28. doi: 10.1038/onc.2015.511. Epub 2016 Feb 8.
|
| 45 |
Ataxia telangiectasia-mutated and p53 are potential mediators of chloroquine-induced resistance to mammary carcinogenesis. Cancer Res. 2007 Dec 15;67(24):12026-33. doi: 10.1158/0008-5472.CAN-07-3058.
|
| 46 |
Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012 Aug 1;131(3):548-57. doi: 10.1002/ijc.26374. Epub 2011 Sep 12.
|
| 47 |
Antitumor activity of chloroquine in combination with Cisplatin in human gastric cancer xenografts. Asian Pac J Cancer Prev. 2015;16(9):3907-12. doi: 10.7314/apjcp.2015.16.9.3907.
|
| 48 |
Critical role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am J Pathol. 2013 Dec;183(6):1815-1825. doi: 10.1016/j.ajpath.2013.08.011. Epub 2013 Oct 1.
|
| 49 |
Riluzole induces AR degradation via endoplasmic reticulum stress pathway in androgen-dependent and castration-resistant prostate cancer cells. Prostate. 2019 Feb;79(2):140-150. doi: 10.1002/pros.23719. Epub 2018 Oct 2.
|
| 50 |
HCV RNA Activates APCs via TLR7/TLR8 While Virus Selectively Stimulates Macrophages Without Inducing Antiviral Responses. Sci Rep. 2016 Jul 7;6:29447. doi: 10.1038/srep29447.
|
| 51 |
Effects of SIDT2 on the miR-25/NOX4/HuR axis and SIRT3 mRNA stability lead to ROS-mediated TNF- expression in hydroquinone-treated leukemia cells. Cell Biol Toxicol. 2023 Oct;39(5):2207-2225. doi: 10.1007/s10565-022-09705-5. Epub 2022 Mar 18.
|
| 52 |
Precipitation of acute intermittent porphyria by chloroquin. Indian Pediatr. 1996 Mar;33(3):241-3.
|
| 53 |
CYP2C8 and antimalaria drug efficacy. Pharmacogenomics. 2007 Feb;8(2):187-98. doi: 10.2217/14622416.8.2.187.
|
| 54 |
NTAL is associated with treatment outcome, cell proliferation and differentiation in acute promyelocytic leukemia. Sci Rep. 2020 Jun 25;10(1):10315. doi: 10.1038/s41598-020-66223-2.
|
| 55 |
A toxicogenomic approach to drug-induced phospholipidosis: analysis of its induction mechanism and establishment of a novel in vitro screening system. Toxicol Sci. 2005 Feb;83(2):282-92.
|
| 56 |
Sertraline induces endoplasmic reticulum stress in hepatic cells. Toxicology. 2014 Aug 1;322:78-88. doi: 10.1016/j.tox.2014.05.007. Epub 2014 May 24.
|
| 57 |
Relationship between release of platelet/endothelial biomarkers and plasma levels of sertraline and N-desmethylsertraline in acute coronary syndrome patients receiving SSRI treatment for depression. Am J Psychiatry. 2005 Jun;162(6):1165-70. doi: 10.1176/appi.ajp.162.6.1165.
|
| 58 |
Antidepressant drug sertraline modulates AMPK-MTOR signaling-mediated autophagy via targeting mitochondrial VDAC1 protein. Autophagy. 2021 Oct;17(10):2783-2799. doi: 10.1080/15548627.2020.1841953. Epub 2020 Nov 9.
|
| 59 |
Psychopharmacological treatment of dermatological patients--when simply talking does not help. J Dtsch Dermatol Ges. 2007 Dec;5(12):1101-6.
|
| 60 |
New orally active anticoagulant agents for the prevention and treatment of venous thromboembolism in cancer patients. Ther Clin Risk Manag. 2014 Jun 13;10:423-36.
|
| 61 |
The selective serotonin reuptake inhibitor sertraline: its profile and use in psychiatric disorders. CNS Drug Rev. 2001 Spring;7(1):1-24.
|
| 62 |
Some aspects of genetic polymorphism in the biotransformation of antidepressants. Therapie. 2004 Jan-Feb;59(1):5-12.
|
| 63 |
PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev Syst Biol Med. 2018 Jul;10(4):e1417. (ID: PA166181117)
|
| 64 |
Sertraline is metabolized by multiple cytochrome P450 enzymes, monoamine oxidases, and glucuronyl transferases in human: an in vitro study. Drug Metab Dispos. 2005 Feb;33(2):262-70.
|
| 65 |
Influence of CYP2B6 and CYP2C19 polymorphisms on sertraline metabolism in major depression patients. Int J Clin Pharm. 2016 Apr;38(2):388-94.
|
| 66 |
Cytochrome P450 inhibition potential of new psychoactive substances of the tryptamine class. Toxicol Lett. 2016 Jan 22;241:82-94.
|
| 67 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
| 68 |
Pharmacokinetics of sertraline in relation to genetic polymorphism of CYP2C19. Clin Pharmacol Ther. 2001 Jul;70(1):42-7.
|
| 69 |
Michaelis-Menten kinetic analysis of drugs of abuse to estimate their affinity to human P-glycoprotein. Toxicol Lett. 2013 Feb 27;217(2):137-42. doi: 10.1016/j.toxlet.2012.12.012. Epub 2012 Dec 27.
|
| 70 |
Immunomodulatory effect of selective serotonin reuptake inhibitors (SSRIs) on human T lymphocyte function and gene expression. Eur Neuropsychopharmacol. 2007 Dec;17(12):774-80.
|
| 71 |
Effects of selective serotonin reuptake inhibitors on three sex steroids in two versions of the aromatase enzyme inhibition assay and in the H295R cell assay. Toxicol In Vitro. 2015 Oct;29(7):1729-35.
|
| 72 |
Sertraline, an antidepressant, induces apoptosis in hepatic cells through the mitogen-activated protein kinase pathway. Toxicol Sci. 2014 Feb;137(2):404-15. doi: 10.1093/toxsci/kft254. Epub 2013 Nov 5.
|
| 73 |
Exploring binding properties of sertraline with human serum albumin: Combination of spectroscopic and molecular modeling studies. Chem Biol Interact. 2015 Dec 5;242:235-46. doi: 10.1016/j.cbi.2015.10.006. Epub 2015 Oct 22.
|
| 74 |
Platelet/endothelial biomarkers in depressed patients treated with the selective serotonin reuptake inhibitor sertraline after acute coronary events: the Sertraline AntiDepressant Heart Attack Randomized Trial (SADHART) Platelet Substudy. Circulation. 2003 Aug 26;108(8):939-44. doi: 10.1161/01.CIR.0000085163.21752.0A. Epub 2003 Aug 11.
|
| 75 |
Impact of selective serotonin reuptake inhibitors on neural crest stem cell formation. Toxicol Lett. 2017 Nov 5;281:20-25. doi: 10.1016/j.toxlet.2017.08.012. Epub 2017 Aug 24.
|
| 76 |
Why are most phospholipidosis inducers also hERG blockers?. Arch Toxicol. 2017 Dec;91(12):3885-3895. doi: 10.1007/s00204-017-1995-9. Epub 2017 May 27.
|
| 77 |
Induction of cysteine string protein after chronic antidepressant treatment in rat frontal cortex. Neurosci Lett. 2001 Apr 6;301(3):183-6. doi: 10.1016/s0304-3940(01)01638-x.
|
| 78 |
The serotonin transporter polymorphism, 5HTTLPR, is associated with a faster response time to sertraline in an elderly population with major depressive disorder. Psychopharmacology (Berl). 2004 Aug;174(4):525-9. doi: 10.1007/s00213-003-1562-3. Epub 2003 Sep 4.
|
|
|
|
|
|
|