| 1 |
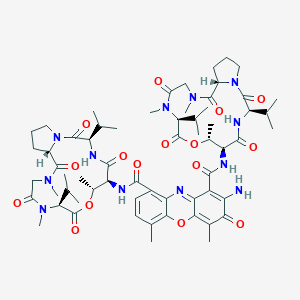
Loss of function mutations in VARS encoding cytoplasmic valyl-tRNA synthetase cause microcephaly, seizures, and progressive cerebral atrophy.Hum Genet. 2018 Apr;137(4):293-303. doi: 10.1007/s00439-018-1882-3. Epub 2018 Apr 24.
|
| 2 |
Dactinomycin FDA Label
|
| 3 |
FDA Approved Drug Products from FDA Official Website. 2009. Application Number: (NDA) 050682.
|
| 4 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 5697).
|
| 5 |
Genomic profiling uncovers a molecular pattern for toxicological characterization of mutagens and promutagens in vitro. Toxicol Sci. 2011 Jul;122(1):185-97.
|
| 6 |
Synergy is achieved by complementation with Apo2L/TRAIL and actinomycin D in Apo2L/TRAIL-mediated apoptosis of prostate cancer cells: role of XIAP in resistance. Prostate. 2002 Dec 1;53(4):286-99. doi: 10.1002/pros.10155.
|
| 7 |
Endocrine disruptors fludioxonil and fenhexamid stimulate miR-21 expression in breast cancer cells. Toxicol Sci. 2013 Jan;131(1):71-83. doi: 10.1093/toxsci/kfs290. Epub 2012 Oct 10.
|
| 8 |
Structural studies of atom-specific anticancer drugs acting on DNA. Pharmacol Ther. 1999 Sep;83(3):181-215.
|
| 9 |
Expression of multidrug resistance-associated protein in NIH/3T3 cells confers multidrug resistance associated with increased drug efflux and altered intracellular drug distribution. Cancer Res. 1995 Nov 15;55(22):5342-7.
|
| 10 |
Potential role of drug transporters in the pathogenesis of medically intractable epilepsy. Epilepsia. 2005 Feb;46(2):224-35.
|
| 11 |
Breast cancer resistance protein (BCRP/ABCG2) induces cellular resistance to HIV-1 nucleoside reverse transcriptase inhibitors. Mol Pharmacol. 2003 Jan;63(1):65-72.
|
| 12 |
Functional analysis of MRP1 cloned from bovine. FEBS Lett. 2002 Jun 19;521(1-3):211-3. doi: 10.1016/s0014-5793(02)02816-8.
|
| 13 |
Establishment and characterization of new cellular lymphoma model expressing transgenic human MDR1. Leuk Res. 2005 Apr;29(4):407-14. doi: 10.1016/j.leukres.2004.09.001.
|
| 14 |
Mechanisms of tissue factor induction by the uremic toxin indole-3 acetic acid through aryl hydrocarbon receptor/nuclear factor-kappa B signaling pathway in human endothelial cells. Arch Toxicol. 2019 Jan;93(1):121-136.
|
| 15 |
Possible roles of a tumor suppressor gene PIG11 in hepatocarcinogenesis and As2O3-induced apoptosis in liver cancer cells. J Gastroenterol. 2009;44(5):460-9. doi: 10.1007/s00535-009-0030-1. Epub 2009 Apr 1.
|
| 16 |
Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J Natl Cancer Inst. 2004 Dec 1;96(23):1769-80. doi: 10.1093/jnci/djh322.
|
| 17 |
Combination of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and actinomycin D induces apoptosis even in TRAIL-resistant human pancreatic cancer cells. Clin Cancer Res. 2001 Feb;7(2):407-14.
|
| 18 |
Response rate of fibrosarcoma cells to cytotoxic drugs on the expression level correlates to the therapeutic response rate of fibrosarcomas and is mediated by regulation of apoptotic pathways. BMC Cancer. 2005 Jul 7;5:74. doi: 10.1186/1471-2407-5-74.
|
| 19 |
Actinomycin D inhibits the expression of the cystine/glutamate transporter xCT via attenuation of CD133 synthesis in CD133(+) HCC. Chem Biol Interact. 2019 Aug 25;309:108713. doi: 10.1016/j.cbi.2019.06.026. Epub 2019 Jun 19.
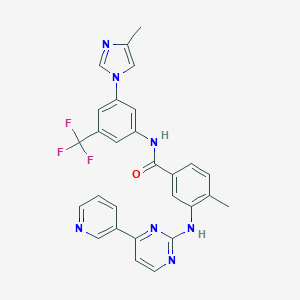
|
| 20 |
Ascorbic acid enhances low-density lipoprotein receptor expression by suppressing proprotein convertase subtilisin/kexin 9 expression. J Biol Chem. 2020 Nov 20;295(47):15870-15882. doi: 10.1074/jbc.RA120.015623. Epub 2020 Sep 10.
|
| 21 |
Heme oxygenase-1-derived carbon monoxide requires the activation of transcription factor NF-kappa B to protect endothelial cells from tumor necrosis factor-alpha-mediated apoptosis. J Biol Chem. 2002 May 17;277(20):17950-61. doi: 10.1074/jbc.M108317200. Epub 2002 Mar 5.
|
| 22 |
The effects of particulate wear debris, cytokines, and growth factors on the functions of MG-63 osteoblasts. J Bone Joint Surg Am. 2001 Feb;83(2):201-11. doi: 10.2106/00004623-200102000-00007.
|
| 23 |
The effect of potent iron chelators on the regulation of p53: examination of the expression, localization and DNA-binding activity of p53 and the transactivation of WAF1. Carcinogenesis. 2003 Oct;24(10):1601-14. doi: 10.1093/carcin/bgg116. Epub 2003 Jul 17.
|
| 24 |
T-2 toxin upregulates the expression of human cytochrome P450 1A1 (CYP1A1) by enhancing NRF1 and Sp1 interaction. Toxicol Lett. 2019 Oct 15;315:77-86. doi: 10.1016/j.toxlet.2019.08.021. Epub 2019 Aug 27.
|
| 25 |
Osmotic Induction of Angiogenic Growth Factor Expression in Human Retinal Pigment Epithelial Cells. PLoS One. 2016 Jan 22;11(1):e0147312. doi: 10.1371/journal.pone.0147312. eCollection 2016.
|
| 26 |
Ammonia promotes endothelial cell survival via the heme oxygenase-1-mediated release of carbon monoxide. Free Radic Biol Med. 2017 Jan;102:37-46. doi: 10.1016/j.freeradbiomed.2016.11.029. Epub 2016 Nov 17.
|
| 27 |
Profiling the immunotoxicity of chemicals based on in vitro evaluation by a combination of the Multi-ImmunoTox assay and the IL-8 Luc assay. Arch Toxicol. 2018 Jun;92(6):2043-2054. doi: 10.1007/s00204-018-2199-7. Epub 2018 Mar 29.
|
| 28 |
Actinomycin D upregulates proapoptotic protein Puma and downregulates Bcl-2 mRNA in normal peripheral blood lymphocytes. Anticancer Drugs. 2007 Aug;18(7):763-72. doi: 10.1097/CAD.0b013e3280adc905.
|
| 29 |
Enhanced GRP78 protein expression via the IRE1/ASK1/p38 MAPK pathway during As(2)O(3)-induced endoplasmic reticulum stress in BEAS-2B cells. Toxicology. 2021 Oct;462:152962. doi: 10.1016/j.tox.2021.152962. Epub 2021 Sep 21.
|
| 30 |
Matrix metalloproteinase-9 is involved in chronic lymphocytic leukemia cell response to fludarabine and arsenic trioxide. PLoS One. 2014 Jun 23;9(6):e99993. doi: 10.1371/journal.pone.0099993. eCollection 2014.
|
| 31 |
Specific inhibition of rRNA transcription and dynamic relocation of fibrillarin induced by mercury. Exp Cell Res. 2000 Aug 25;259(1):225-38. doi: 10.1006/excr.2000.4923.
|
| 32 |
Phosphorylation of the RNA polymerase II largest subunit during heat shock and inhibition of transcription in HeLa cells. J Cell Physiol. 1994 Mar;158(3):417-26. doi: 10.1002/jcp.1041580305.
|
| 33 |
Selective and nonselective toxicity of TRAIL/Apo2L combined with chemotherapy in human bone tumour cells vs. normal human cells. Int J Cancer. 2003 Dec 20;107(6):929-40. doi: 10.1002/ijc.11503.
|
| 34 |
Actinomycin D and staurosporine, potent apoptosis inducers in vitro, are potentially effective chemotherapeutic agents against glioblastoma multiforme. Cancer Chemother Pharmacol. 2000;45(2):149-56. doi: 10.1007/s002800050023.
|
| 35 |
Unpredicted Downregulation of RAD51 Suggests Genome Instability Induced by Tetrachlorobenzoquinone. Chem Res Toxicol. 2016 Dec 19;29(12):2184-2193. doi: 10.1021/acs.chemrestox.6b00369. Epub 2016 Nov 21.
|
| 36 |
Mechanisms of thymidine kinase/ganciclovir and cytosine deaminase/ 5-fluorocytosine suicide gene therapy-induced cell death in glioma cells. Oncogene. 2005 Feb 10;24(7):1231-43. doi: 10.1038/sj.onc.1208290.
|
| 37 |
Quercetin downregulates Mcl-1 by acting on mRNA stability and protein degradation. Br J Cancer. 2011 Jul 12;105(2):221-30. doi: 10.1038/bjc.2011.229.
|
| 38 |
Androgen receptor-dependent transactivation of growth arrest-specific gene 6 mediates inhibitory effects of testosterone on vascular calcification. J Biol Chem. 2010 Mar 5;285(10):7537-44. doi: 10.1074/jbc.M109.055087. Epub 2010 Jan 4.
|
| 39 |
A high-throughput screen for teratogens using human pluripotent stem cells. Toxicol Sci. 2014 Jan;137(1):76-90. doi: 10.1093/toxsci/kft239. Epub 2013 Oct 23.
|
| 40 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
| 41 |
[Expression of C1QBP gene and its correlation with drug resistance in human resistance choriocarcinoma cell line]. Zhonghua Fu Chan Ke Za Zhi. 2014 Aug;49(8):616-20.
|
| 42 |
Assessment of the inhibition potential of Licochalcone A against human UDP-glucuronosyltransferases. Food Chem Toxicol. 2016 Apr;90:112-22.
|
| 43 |
Exposure-based assessment of chemical teratogenicity using morphogenetic aggregates of human embryonic stem cells. Reprod Toxicol. 2020 Jan;91:74-91. doi: 10.1016/j.reprotox.2019.10.004. Epub 2019 Nov 8.
|
| 44 |
Endoplasmic reticulum stress-mediated apoptosis in imatinib-resistant leukemic K562-r cells triggered by AMN107 combined with arsenic trioxide. Exp Biol Med (Maywood). 2013 Aug 1;238(8):932-42. doi: 10.1177/1535370213492689. Epub 2013 Jul 24.
|
| 45 |
2007 FDA drug approvals: a year of flux. Nat Rev Drug Discov. 2008 Feb;7(2):107-9.
|
| 46 |
Interaction of nilotinib, dasatinib and bosutinib with ABCB1 and ABCG2: implications for altered anti-cancer effects and pharmacological properties. Br J Pharmacol. 2009 Oct;158(4):1153-64.
|
| 47 |
KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017 Jan 4;45(D1):D353-D361. (dg:DG01665)
|
| 48 |
Contribution of OATP1B1 and OATP1B3 to the disposition of sorafenib and sorafenib-glucuronide. Clin Cancer Res. 2013 Mar 15;19(6):1458-66.
|
| 49 |
Drug interactions with the tyrosine kinase inhibitors imatinib, dasatinib, and nilotinib. Blood. 2011 Feb 24;117(8):e75-87.
|
| 50 |
Role of cytochrome P450 2C8 in drug metabolism and interactions. Pharmacol Rev. 2016 Jan;68(1):168-241.
|
| 51 |
Resistance to daunorubicin, imatinib, or nilotinib depends on expression levels of ABCB1 and ABCG2 in human leukemia cells. Chem Biol Interact. 2014 Aug 5;219:203-10. doi: 10.1016/j.cbi.2014.06.009. Epub 2014 Jun 19.
|
| 52 |
Reversal of ABCB1 mediated efflux by imatinib and nilotinib in cells expressing various transporter levels. Chem Biol Interact. 2017 Aug 1;273:171-179. doi: 10.1016/j.cbi.2017.06.012. Epub 2017 Jun 13.
|
| 53 |
Multi-parameter in vitro toxicity testing of crizotinib, sunitinib, erlotinib, and nilotinib in human cardiomyocytes. Toxicol Appl Pharmacol. 2013 Oct 1;272(1):245-55.
|
| 54 |
p53 Gene (NY-CO-13) Levels in Patients with Chronic Myeloid Leukemia: The Role of Imatinib and Nilotinib. Diseases. 2018 Jan 25;6(1):13. doi: 10.3390/diseases6010013.
|
| 55 |
Nilotinib reduced the viability of human ovarian cancer cells via mitochondria-dependent apoptosis, independent of JNK activation. Toxicol In Vitro. 2016 Mar;31:1-11. doi: 10.1016/j.tiv.2015.11.002. Epub 2015 Nov 6.
|
| 56 |
AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009 Nov 6;16(5):401-12. doi: 10.1016/j.ccr.2009.09.028.
|
| 57 |
The CML-related oncoprotein BCR/ABL induces expression of histidine decarboxylase (HDC) and the synthesis of histamine in leukemic cells. Blood. 2006 Nov 15;108(10):3538-47. doi: 10.1182/blood-2005-12-028456. Epub 2006 Jul 18.
|
| 58 |
Cytotoxicity of 34 FDA approved small-molecule kinase inhibitors in primary rat and human hepatocytes. Toxicol Lett. 2018 Jul;291:138-148. doi: 10.1016/j.toxlet.2018.04.010. Epub 2018 Apr 12.
|
| 59 |
Biologically active neutrophil chemokine pattern in tonsillitis.Clin Exp Immunol. 2004 Mar;135(3):511-8. doi: 10.1111/j.1365-2249.2003.02390.x.
|
|
|
|
|
|
|