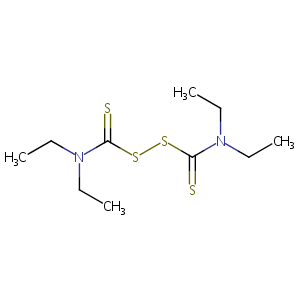
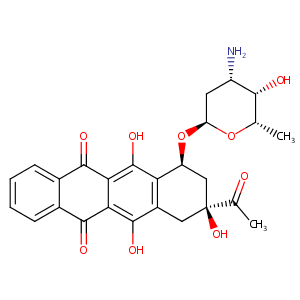
| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015
|
| 3 |
Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020 Apr 9.
|
| 4 |
Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov. 2020 Mar;19(3):149-150.
|
| 5 |
Idarubicin FDA Label
|
| 6 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7083).
|
| 7 |
Keratinocyte gene expression profiles discriminate sensitizing and irritating compounds. Toxicol Sci. 2010 Sep;117(1):81-9.
|
| 8 |
Pharmacological treatment of alcohol dependence: target symptoms and target mechanisms. Pharmacol Ther. 2006 Sep;111(3):855-76.
|
| 9 |
Interaction of disulfiram with antiretroviral medications: efavirenz increases while atazanavir decreases disulfiram effect on enzymes of alcohol metabolism. Am J Addict. 2014 Mar-Apr;23(2):137-44.
|
| 10 |
Identification of the human P-450 enzymes responsible for the sulfoxidation and thiono-oxidation of diethyldithiocarbamate methyl ester: role of P-450 enzymes in disulfiram bioactivation. Alcohol Clin Exp Res. 1998 Sep;22(6):1212-9.
|
| 11 |
Duration of cytochrome P-450 2E1 (CYP2E1) inhibition and estimation of functional CYP2E1 enzyme half-life after single-dose disulfiram administration in humans. J Pharmacol Exp Ther. 1999 Oct;291(1):213-9.
|
| 12 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
| 13 |
Hypotension with anesthesia in disulfiram-treated patients. Anesthesiology. 1979 Oct;51(4):366-8.
|
| 14 |
Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol. 1991 Mar-Apr;4(2):168-79.
|
| 15 |
The molecular basis of the action of disulfiram as a modulator of the multidrug resistance-linked ATP binding cassette transporters MDR1 (ABCB1) and MRP1 (ABCC1). Mol Pharmacol. 2004 Mar;65(3):675-84.
|
| 16 |
High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin Cancer Res. 2009 Oct 1;15(19):6070-8.
|
| 17 |
The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact. 2012 Jan 5;195(1):52-60.
|
| 18 |
Species-specific differences in the inhibition of human and zebrafish 11beta-hydroxysteroid dehydrogenase 2 by thiram and organotins. Toxicology. 2012 Nov 15;301(1-3):72-8.
|
| 19 |
Early identification of clinically relevant drug interactions with the human bile salt export pump (BSEP/ABCB11). Toxicol Sci. 2013 Dec;136(2):328-43.
|
| 20 |
The interaction of disulfiram and H(2)S metabolism in inhibition of aldehyde dehydrogenase activity and liver cancer cell growth. Toxicol Appl Pharmacol. 2021 Sep 1;426:115642. doi: 10.1016/j.taap.2021.115642. Epub 2021 Jul 6.
|
| 21 |
Identification and Profiling of Environmental Chemicals That Inhibit the TGF/SMAD Signaling Pathway. Chem Res Toxicol. 2019 Dec 16;32(12):2433-2444. doi: 10.1021/acs.chemrestox.9b00228. Epub 2019 Nov 11.
|
| 22 |
Comparative study of the effects of ziram and disulfiram on human monocyte-derived macrophage functions and polarization: involvement of zinc. Cell Biol Toxicol. 2021 Jun;37(3):379-400. doi: 10.1007/s10565-020-09540-6. Epub 2020 Jul 25.
|
| 23 |
Association of CYP1A1 and CYP1B1 inhibition in in vitro assays with drug-induced liver injury. J Toxicol Sci. 2021;46(4):167-176. doi: 10.2131/jts.46.167.
|
| 24 |
Disulfiram metabolites permanently inactivate the human multidrug resistance P-glycoprotein. Mol Pharm. 2004 Nov-Dec;1(6):426-33. doi: 10.1021/mp049917l.
|
| 25 |
Disulfiram suppresses invasive ability of osteosarcoma cells via the inhibition of MMP-2 and MMP-9 expression. J Biochem Mol Biol. 2007 Nov 30;40(6):1069-76. doi: 10.5483/bmbrep.2007.40.6.1069.
|
| 26 |
Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006 Nov 1;66(21):10425-33. doi: 10.1158/0008-5472.CAN-06-2126.
|
| 27 |
Low-Dose Pesticides Alter Primary Human Bone Marrow Mesenchymal Stem/Stromal Cells through ALDH2 Inhibition. Cancers (Basel). 2021 Nov 14;13(22):5699. doi: 10.3390/cancers13225699.
|
| 28 |
Disulfiram is a direct and potent inhibitor of human O6-methylguanine-DNA methyltransferase (MGMT) in brain tumor cells and mouse brain and markedly increases the alkylating DNA damage. Carcinogenesis. 2014 Mar;35(3):692-702. doi: 10.1093/carcin/bgt366. Epub 2013 Nov 5.
|
| 29 |
Identification of novel activators of the metal responsive transcription factor (MTF-1) using a gene expression biomarker in a microarray compendium. Metallomics. 2020 Sep 23;12(9):1400-1415. doi: 10.1039/d0mt00071j.
|
| 30 |
Pharmacological profiling of disulfiram using human tumor cell lines and human tumor cells from patients. Biochem Pharmacol. 2007 Jan 1;73(1):25-33. doi: 10.1016/j.bcp.2006.08.016. Epub 2006 Aug 26.
|
| 31 |
Disulfiram inhibits inflammation and fibrosis in a rat unilateral ureteral obstruction model by inhibiting gasdermin D cleavage and pyroptosis. Inflamm Res. 2021 May;70(5):543-552. doi: 10.1007/s00011-021-01457-y. Epub 2021 Apr 13.
|
| 32 |
Cytochrome P(450)-dependent toxic effects of primaquine on human erythrocytes. Toxicol Appl Pharmacol. 2009 Nov 15;241(1):14-22. doi: 10.1016/j.taap.2009.07.012. Epub 2009 Jul 17.
|
| 33 |
Effect of common medications on the expression of SARS-CoV-2 entry receptors in liver tissue. Arch Toxicol. 2020 Dec;94(12):4037-4041. doi: 10.1007/s00204-020-02869-1. Epub 2020 Aug 17.
|
| 34 |
Contribution of aldehyde dehydrogenase 3A1 to disulfiram penetration through monolayers consisting of cultured human corneal epithelial cells. Biol Pharm Bull. 2008 Jul;31(7):1444-8. doi: 10.1248/bpb.31.1444.
|
| 35 |
Disulfiram facilitates ataxin-3 nuclear translocation and potentiates the cytotoxicity in a cell model of SCA3. J Toxicol Sci. 2019;44(8):535-542. doi: 10.2131/jts.44.535.
|
| 36 |
Development of a highly sensitive cytotoxicity assay system for CYP3A4-mediated metabolic activation. Drug Metab Dispos. 2011 Aug;39(8):1388-95. doi: 10.1124/dmd.110.037077. Epub 2011 May 3.
|
| 37 |
Quantitative high-throughput profiling of environmental chemicals and drugs that modulate farnesoid X receptor. Sci Rep. 2014 Sep 26;4:6437. doi: 10.1038/srep06437.
|
| 38 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services.
|
| 39 |
Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions. Pharm Res. 2013 Apr;30(4):996-1007.
|
| 40 |
Amonafide L-malate is not a substrate for multidrug resistance proteins in secondary acute myeloid leukemia. Leukemia. 2008 Nov;22(11):2110-5.
|
| 41 |
In vitro evaluation of cytochrome P450-mediated drug interactions between cytarabine, idarubicin, itraconazole and caspofungin. Hematology. 2004 Jun;9(3):217-21.
|
| 42 |
A Quantitative Approach to Screen for Nephrotoxic Compounds In Vitro. J Am Soc Nephrol. 2016 Apr;27(4):1015-28. doi: 10.1681/ASN.2015010060. Epub 2015 Aug 10.
|
| 43 |
The use of biochemical markers in cardiotoxicity monitoring in patients treated for leukemia. Neoplasma. 2005;52(5):430-4.
|
| 44 |
The induction of apoptosis by daunorubicin and idarubicin in human trisomic and diabetic fibroblasts. Cell Mol Biol Lett. 2008;13(2):182-94. doi: 10.2478/s11658-007-0045-7. Epub 2008 Apr 10.
|
| 45 |
Refining the human iPSC-cardiomyocyte arrhythmic risk assessment model. Toxicol Sci. 2013 Dec;136(2):581-94. doi: 10.1093/toxsci/kft205. Epub 2013 Sep 19.
|
|
|
|
|
|
|