| 1 |
Recurrent recessive mutation in deoxyguanosine kinase causes idiopathic noncirrhotic portal hypertension.Hepatology. 2016 Jun;63(6):1977-86. doi: 10.1002/hep.28499. Epub 2016 Mar 31.
|
| 2 |
Natural products as sources of new drugs over the last 25 years. J Nat Prod. 2007 Mar;70(3):461-77.
|
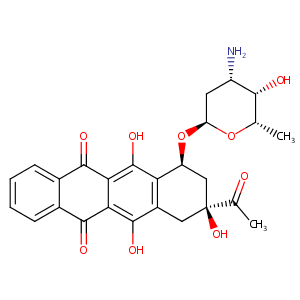
| 3 |
Idarubicin FDA Label
|
| 4 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 7083).
|
| 5 |
A potent HIV protease inhibitor, darunavir, does not inhibit ZMPSTE24 or lead to an accumulation of farnesyl-prelamin A in cells. J Biol Chem. 2008 Apr 11;283(15):9797-804. doi: 10.1074/jbc.M709629200. Epub 2008 Jan 28.
|
| 6 |
Modulation of UDP-glucuronosyltransferase 1A1 in primary human hepatocytes by prototypical inducers. J Biochem Mol Toxicol. 2005;19(2):96-108. doi: 10.1002/jbt.20058.
|
| 7 |
Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int J Antimicrob Agents. 2009 Apr;33(4):307-20.
|
| 8 |
Interaction of eight HIV protease inhibitors with the canalicular efflux transporter ABCC2 (MRP2) in sandwich-cultured rat and human hepatocytes. Biopharm Drug Dispos. 2010 Mar;31(2-3):178-88.
|
| 9 |
MDR1 G1199A polymorphism alters permeability of HIV protease inhibitors across P-glycoprotein-expressing epithelial cells. AIDS. 2005 Oct 14;19(15):1617-25.
|
| 10 |
pH dependence of organic anion-transporting polypeptide 2B1 in Caco-2 cells: potential role in antiretroviral drug oral bioavailability and drug-drug interactions. J Pharmacol Exp Ther. 2010 Sep 1;334(3):1009-22.
|
| 11 |
Effect of extended exposure to grapefruit juice on cytochrome P450 3A activity in humans: comparison with ritonavir. Clin Pharmacol Ther. 2006 Mar;79(3):243-54.
|
| 12 |
Reversible coma caused by risperidone-ritonavir interaction. Clin Neuropharmacol. 2002 Sep-Oct;25(5):251-3.
|
| 13 |
Drug Interactions Flockhart Table
|
| 14 |
Evaluation of CYP2B6 induction and prediction of clinical drug-drug interactions: considerations from the IQ consortium induction working group-an industry perspective. Drug Metab Dispos. 2016 Oct;44(10):1720-30.
|
| 15 |
Mechanism-based inactivation of CYP3A by HIV protease inhibitors. J Pharmacol Exp Ther. 2005 Feb;312(2):583-91.
|
| 16 |
Transcriptional profiling suggests that Nevirapine and Ritonavir cause drug induced liver injury through distinct mechanisms in primary human hepatocytes. Chem Biol Interact. 2016 Aug 5;255:31-44.
|
| 17 |
Use of immortalized human hepatocytes to predict the magnitude of clinical drug-drug interactions caused by CYP3A4 induction. Drug Metab Dispos. 2006 Oct;34(10):1742-8.
|
| 18 |
Some HIV antiretrovirals increase oxidative stress and alter chemokine, cytokine or adiponectin production in human adipocytes and macrophages. Antivir Ther. 2007;12(4):489-500.
|
| 19 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
| 20 |
Severe interaction between ritonavir and acenocoumarol. Ann Pharmacother. 2002 Apr;36(4):621-3.
|
| 21 |
Examination of 209 drugs for inhibition of cytochrome P450 2C8. J Clin Pharmacol. 2005 Jan;45(1):68-78.
|
| 22 |
Interference with bile salt export pump function is a susceptibility factor for human liver injury in drug development. Toxicol Sci. 2010 Dec; 118(2):485-500.
|
| 23 |
Ritonavir blocks AKT signaling, activates apoptosis and inhibits migration and invasion in ovarian cancer cells. Mol Cancer. 2009 Apr 22;8:26. doi: 10.1186/1476-4598-8-26.
|
| 24 |
Efficient intervention of growth and infiltration of primary adult T-cell leukemia cells by an HIV protease inhibitor, ritonavir. Blood. 2006 Jan 15;107(2):716-24. doi: 10.1182/blood-2005-02-0735. Epub 2005 Sep 20.
|
| 25 |
The HIV protease inhibitor ritonavir synergizes with butyrate for induction of apoptotic cell death and mediates expression of heme oxygenase-1 in DLD-1 colon carcinoma cells. Br J Pharmacol. 2004 Dec;143(7):890-8. doi: 10.1038/sj.bjp.0706023. Epub 2004 Oct 25.
|
| 26 |
HIV-1 protease inhibitor ritonavir modulates susceptibility to apoptosis of uninfected T cells. J Hum Virol. 1999 Sep-Oct;2(5):261-9.
|
| 27 |
Premature senescence of vascular cells is induced by HIV protease inhibitors: implication of prelamin A and reversion by statin. Arterioscler Thromb Vasc Biol. 2010 Dec;30(12):2611-20. doi: 10.1161/ATVBAHA.110.213603. Epub 2010 Sep 30.
|
| 28 |
Binding of anti-HIV drugs to human serum albumin. IUBMB Life. 2004 Oct;56(10):609-14. doi: 10.1080/15216540400016286.
|
| 29 |
Effects of HIV protease inhibitor ritonavir on Akt-regulated cell proliferation in breast cancer. Clin Cancer Res. 2006 Mar 15;12(6):1883-96. doi: 10.1158/1078-0432.CCR-05-1167.
|
| 30 |
Heme oxygenase-1-derived bilirubin counteracts HIV protease inhibitor-mediated endothelial cell dysfunction. Free Radic Biol Med. 2016 May;94:218-29. doi: 10.1016/j.freeradbiomed.2016.03.003. Epub 2016 Mar 8.
|
| 31 |
The human immunodeficiency virus protease inhibitor ritonavir inhibits lung cancer cells, in part, by inhibition of survivin. J Thorac Oncol. 2011 Apr;6(4):661-70. doi: 10.1097/JTO.0b013e31820c9e3c.
|
| 32 |
Evaluating the Role of Multidrug Resistance Protein 3 (MDR3) Inhibition in Predicting Drug-Induced Liver Injury Using 125 Pharmaceuticals. Chem Res Toxicol. 2017 May 15;30(5):1219-1229. doi: 10.1021/acs.chemrestox.7b00048. Epub 2017 May 4.
|
| 33 |
Nordihydroguaiaretic acid (NDGA) inhibits ritonavir-induced endothelial dysfunction in porcine pulmonary arteries. Med Sci Monit. 2011 Nov;17(11):BR312-318. doi: 10.12659/msm.882040.
|
| 34 |
Estrogen prevents cholesteryl ester accumulation in macrophages induced by the HIV protease inhibitor ritonavir. J Cell Biochem. 2008 Apr 1;103(5):1598-606. doi: 10.1002/jcb.21546.
|
| 35 |
Association of CYP1A1 and CYP1B1 inhibition in in vitro assays with drug-induced liver injury. J Toxicol Sci. 2021;46(4):167-176. doi: 10.2131/jts.46.167.
|
| 36 |
Quantitative high-throughput profiling of environmental chemicals and drugs that modulate farnesoid X receptor. Sci Rep. 2014 Sep 26;4:6437. doi: 10.1038/srep06437.
|
| 37 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services.
|
| 38 |
Human intestinal transporter database: QSAR modeling and virtual profiling of drug uptake, efflux and interactions. Pharm Res. 2013 Apr;30(4):996-1007.
|
| 39 |
Amonafide L-malate is not a substrate for multidrug resistance proteins in secondary acute myeloid leukemia. Leukemia. 2008 Nov;22(11):2110-5.
|
| 40 |
In vitro evaluation of cytochrome P450-mediated drug interactions between cytarabine, idarubicin, itraconazole and caspofungin. Hematology. 2004 Jun;9(3):217-21.
|
| 41 |
A Quantitative Approach to Screen for Nephrotoxic Compounds In Vitro. J Am Soc Nephrol. 2016 Apr;27(4):1015-28. doi: 10.1681/ASN.2015010060. Epub 2015 Aug 10.
|
| 42 |
The use of biochemical markers in cardiotoxicity monitoring in patients treated for leukemia. Neoplasma. 2005;52(5):430-4.
|
| 43 |
The induction of apoptosis by daunorubicin and idarubicin in human trisomic and diabetic fibroblasts. Cell Mol Biol Lett. 2008;13(2):182-94. doi: 10.2478/s11658-007-0045-7. Epub 2008 Apr 10.
|
| 44 |
Refining the human iPSC-cardiomyocyte arrhythmic risk assessment model. Toxicol Sci. 2013 Dec;136(2):581-94. doi: 10.1093/toxsci/kft205. Epub 2013 Sep 19.
|
|
|
|
|
|
|