| 1 |
ClinicalTrials.gov (NCT05792540) Clinical Study to Evaluate the Possible Efficacy of Dapagliflozin and Atorvastatin in Patients With Major Depressive Disorders
|
| 2 |
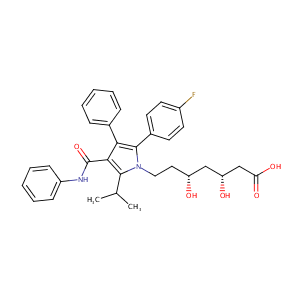
Atorvastatin FDA Label
|
| 3 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015
|
| 4 |
FDA Approved Drug Products from FDA Official Website. 2019. Application Number: (ANDA) 205945.
|
| 5 |
ClinicalTrials.gov (NCT04380402) Atorvastatin in COVID-19. U.S. National Institutes of Health.
|
| 6 |
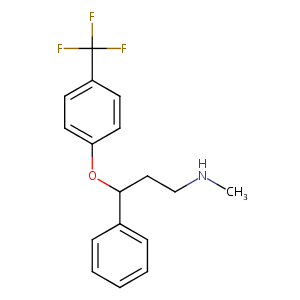
Fluoxetine FDA Label
|
| 7 |
URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Ligand id: 203).
|
| 8 |
Fluoxetine to Reduce Intubation and Death After COVID19 Infection
|
| 9 |
Atorvastatin affects several angiogenic mediators in human endothelial cells. Endothelium. 2005 Sep-Dec;12(5-6):233-41.
|
| 10 |
Markers of inflammation, thrombosis and endothelial activation correlate with carotid IMT regression in stable coronary disease after atorvastatin treatment. Nutr Metab Cardiovasc Dis. 2009 Sep;19(7):481-90. doi: 10.1016/j.numecd.2008.10.003. Epub 2009 Jan 26.
|
| 11 |
Simvastatin reduces VCAM-1 expression in human umbilical vein endothelial cells exposed to lipopolysaccharide. Inflamm Res. 2012 May;61(5):485-91. doi: 10.1007/s00011-012-0435-9. Epub 2012 Jan 15.
|
| 12 |
Equally potent inhibitors of cholesterol synthesis in human hepatocytes have distinguishable effects on different cytochrome P450 enzymes. Biopharm Drug Dispos. 2000 Dec;21(9):353-64.
|
| 13 |
Statin therapy in COVID-19 infection. Eur Heart J Cardiovasc Pharmacother. 2020 Apr 29. pii: pvaa042.
|
| 14 |
Tarascon Pocket Pharmacopoeia 2018 Classic Shirt-Pocket Edition.
|
| 15 |
Rifampicin alters atorvastatin plasma concentration on the basis of SLCO1B1 521T>C polymorphism. Clin Chim Acta. 2009 Jul;405(1-2):49-52.
|
| 16 |
Influence of the flavonoids apigenin, kaempferol, and quercetin on the function of organic anion transporting polypeptides 1A2 and 2B1. Biochem Pharmacol. 2010 Dec 1;80(11):1746-53.
|
| 17 |
FDA Drug Development and Drug Interactions
|
| 18 |
Human platelets express organic anion-transporting peptide 2B1, an uptake transporter for atorvastatin. Drug Metab Dispos. 2009 May;37(5):1129-37.
|
| 19 |
H+-coupled nutrient, micronutrient and drug transporters in the mammalian small intestine. Exp Physiol. 2007 Jul;92(4):603-19.
|
| 20 |
Monocarboxylate Transporters in Drug Disposition: Role in the Toxicokinetics and Toxicodynamics of the Drug of Abuse GHB.
|
| 21 |
Substrates, inducers, inhibitors and structure-activity relationships of human Cytochrome P450 2C9 and implications in drug development. Curr Med Chem. 2009;16(27):3480-675.
|
| 22 |
Atorvastatin glucuronidation is minimally and nonselectively inhibited by the fibrates gemfibrozil, fenofibrate, and fenofibric acid. Drug Metab Dispos. 2007 Aug;35(8):1315-24.
|
| 23 |
Effects of acid and lactone forms of eight HMG-CoA reductase inhibitors on CYP-mediated metabolism and MDR1-mediated transport. Pharm Res. 2006 Mar;23(3):506-12.
|
| 24 |
Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers.
|
| 25 |
Role of cytochrome P450 2C8 in drug metabolism and interactions. Pharmacol Rev. 2016 Jan;68(1):168-241.
|
| 26 |
UGT1A1*28 is associated with decreased systemic exposure of atorvastatin lactone. Mol Diagn Ther. 2013 Aug;17(4):233-7.
|
| 27 |
ADReCS-Target: target profiles for aiding drug safety research and application. Nucleic Acids Res. 2018 Jan 4;46(D1):D911-D917. doi: 10.1093/nar/gkx899.
|
| 28 |
Effect of atorvastatin and fluvastatin on the metabolism of midazolam by cytochrome P450 in vitro. Anaesthesia. 2005 Aug;60(8):747-53.
|
| 29 |
Effects of rosiglitazone and atorvastatin on the expression of genes that control cholesterol homeostasis in differentiating monocytes. Biochem Pharmacol. 2006 Feb 28;71(5):605-14.
|
| 30 |
Differential regulation of apolipoprotein B secretion from HepG2 cells by two HMG-CoA reductase inhibitors, atorvastatin and simvastatin. J Lipid Res. 1999 Jun;40(6):1078-89.
|
| 31 |
[Association of a polymorphic marker Trp719Arg of KIF6 gene with effects of atorvastatin and simvastatin in patients with early ischemic heart disease]. Kardiologiia. 2011;51(9):4-12.
|
| 32 |
Differential interaction of 3-hydroxy-3-methylglutaryl-coa reductase inhibitors with ABCB1, ABCC2, and OATP1B1. Drug Metab Dispos. 2005 Apr;33(4):537-46.
|
| 33 |
Effect of pitavastatin on transactivation of human serum paraoxonase 1 gene. Metabolism. 2005 Feb;54(2):142-50.
|
| 34 |
Identification of HMG-CoA reductase inhibitors as activators for human, mouse and rat constitutive androstane receptor. Drug Metab Dispos. 2005 Jul;33(7):924-9.
|
| 35 |
The effect of statins on lipids peroxidation and activities of antioxidants enzymes in patients with dyslipidemia. Przegl Lek. 2006;63(9):738-42.
|
| 36 |
A multifactorial approach to hepatobiliary transporter assessment enables improved therapeutic compound development. Toxicol Sci. 2013 Nov;136(1):216-41.
|
| 37 |
Involvement of pregnane X receptor in the impaired glucose utilization induced by atorvastatin in hepatocytes. Biochem Pharmacol. 2016 Jan 15;100:98-111.
|
| 38 |
Differential effects of statins on relevant functions of human monocyte-derived dendritic cells. J Leukoc Biol. 2006 Mar;79(3):529-38. doi: 10.1189/jlb.0205064. Epub 2005 Dec 30.
|
| 39 |
Atorvastatin and quinapril inhibit blood coagulation in patients with coronary artery disease following 28 days of therapy. J Thromb Haemost. 2006 Nov;4(11):2397-404. doi: 10.1111/j.1538-7836.2006.02165.x. Epub 2006 Aug 14.
|
| 40 |
Atorvastatin induces MicroRNA-145 expression in HEPG2 cells via regulation of the PI3K/AKT signalling pathway. Chem Biol Interact. 2018 May 1;287:32-40. doi: 10.1016/j.cbi.2018.04.005. Epub 2018 Apr 6.
|
| 41 |
Additive beneficial effects of atorvastatin combined with amlodipine in patients with mild-to-moderate hypertension. Int J Cardiol. 2011 Feb 3;146(3):319-25. doi: 10.1016/j.ijcard.2009.07.002. Epub 2009 Aug 3.
|
| 42 |
Simvastatin and atorvastatin enhance gene expression of collagen type 1 and osteocalcin in primary human osteoblasts and MG-63 cultures. J Cell Biochem. 2007 Aug 15;101(6):1430-8. doi: 10.1002/jcb.21259.
|
| 43 |
Effects of atorvastatin on inflammation and oxidative stress. Heart Vessels. 2005 Jul;20(4):133-6. doi: 10.1007/s00380-005-0833-9.
|
| 44 |
Use of atorvastatin in hyperlipidemic hypertensive renal transplant recipients. Pediatr Nephrol. 2002 Jul;17(7):540-3. doi: 10.1007/s00467-002-0860-z. Epub 2002 May 25.
|
| 45 |
Atorvastatin induces mitochondrial dysfunction and cell apoptosis in HepG2 cells via inhibition of the Nrf2 pathway. J Appl Toxicol. 2019 Oct;39(10):1394-1404. doi: 10.1002/jat.3825. Epub 2019 Aug 18.
|
| 46 |
Inhibition of autophagy enhances anticancer effects of atorvastatin in digestive malignancies. Cancer Res. 2010 Oct 1;70(19):7699-709. doi: 10.1158/0008-5472.CAN-10-1626. Epub 2010 Sep 28.
|
| 47 |
Simvastatin reduces expression of cytokines interleukin-6, interleukin-8, and monocyte chemoattractant protein-1 in circulating monocytes from hypercholesterolemic patients. Arterioscler Thromb Vasc Biol. 2002 Jul 1;22(7):1194-9. doi: 10.1161/01.atv.0000022694.16328.cc.
|
| 48 |
The effects of atorvastatin (10 mg) on systemic inflammation in heart failure. Am J Cardiol. 2005 Dec 15;96(12):1699-704. doi: 10.1016/j.amjcard.2005.07.092. Epub 2005 Oct 28.
|
| 49 |
Statins regulate alpha2beta1-integrin expression and collagen I-dependent functions in human vascular smooth muscle cells. J Cardiovasc Pharmacol. 2003 Jan;41(1):89-96. doi: 10.1097/00005344-200301000-00012.
|
| 50 |
Combination of atorvastatin and celecoxib synergistically induces cell cycle arrest and apoptosis in colon cancer cells. Int J Cancer. 2008 May 1;122(9):2115-24. doi: 10.1002/ijc.23315.
|
| 51 |
Integrin expression on monocytes and lymphocytes in unstable angina short term effects of atorvastatin. Rom J Intern Med. 2007;45(2):193-9.
|
| 52 |
RhoA GTPase inactivation by statins induces osteosarcoma cell apoptosis by inhibiting p42/p44-MAPKs-Bcl-2 signaling independently of BMP-2 and cell differentiation. Cell Death Differ. 2006 Nov;13(11):1845-56. doi: 10.1038/sj.cdd.4401873. Epub 2006 Feb 10.
|
| 53 |
Treatment with atorvastatin alters the ratio of interleukin-12/interleukin-10 gene expression [corrected]. Eur J Clin Invest. 2003 Jan;33(1):88-91. doi: 10.1046/j.1365-2362.2003.01105.x.
|
| 54 |
Effect of atorvastatin on circulating proinflammatory T-lymphocyte subsets and soluble CD40 ligand in patients with stable coronary artery disease--a randomized, placebo-controlled study. Am Heart J. 2006 Jan;151(1):139. doi: 10.1016/j.ahj.2005.10.006.
|
| 55 |
Atorvastatin and pravastatin stimulate nitric oxide and reactive oxygen species generation, affect mitochondrial network architecture and elevate nicotinamide N-methyltransferase level in endothelial cells. J Appl Toxicol. 2021 Jul;41(7):1076-1088. doi: 10.1002/jat.4094. Epub 2020 Oct 19.
|
| 56 |
Decreased circulating Fas ligand in patients with familial combined hyperlipidemia or carotid atherosclerosis: normalization by atorvastatin. J Am Coll Cardiol. 2004 Apr 7;43(7):1188-94. doi: 10.1016/j.jacc.2003.10.046.
|
| 57 |
Effect of atorvastatin, simvastatin, and lovastatin on the metabolism of cholesterol and triacylglycerides in HepG2 cells. Biochem Pharmacol. 2001 Dec 1;62(11):1545-55. doi: 10.1016/s0006-2952(01)00790-0.
|
| 58 |
Effect of Ivermectin and Atorvastatin on Nuclear Localization of Importin Alpha and Drug Target Expression Profiling in Host Cells from Nasopharyngeal Swabs of SARS-CoV-2- Positive Patients. Viruses. 2021 Oct 15;13(10):2084. doi: 10.3390/v13102084.
|
| 59 |
Statins activate the mitochondrial pathway of apoptosis in human lymphoblasts and myeloma cells. Carcinogenesis. 2005 May;26(5):883-91. doi: 10.1093/carcin/bgi036. Epub 2005 Feb 10.
|
| 60 |
Atorvastatin mediates increases in intralesional BAX and BAK expression in human end-stage abdominal aortic aneurysms. Can J Physiol Pharmacol. 2009 Nov;87(11):915-22. doi: 10.1139/y09-085.
|
| 61 |
Regulation mechanism of ABCA1 expression by statins in hepatocytes. Eur J Pharmacol. 2011 Jul 15;662(1-3):9-14. doi: 10.1016/j.ejphar.2011.04.043. Epub 2011 May 1.
|
| 62 |
Decreased macrophage paraoxonase 2 expression in patients with hypercholesterolemia is the result of their increased cellular cholesterol content: effect of atorvastatin therapy. Arterioscler Thromb Vasc Biol. 2004 Jan;24(1):175-80. doi: 10.1161/01.ATV.0000104011.88939.06. Epub 2003 Oct 30.
|
| 63 |
Regulation of apoptosis in human melanoma and neuroblastoma cells by statins, sodium arsenite and TRAIL: a role of combined treatment versus monotherapy. Apoptosis. 2011 Dec;16(12):1268-84.
|
| 64 |
Atorvastatin transport in the Caco-2 cell model: contributions of P-glycoprotein and the proton-monocarboxylic acid co-transporter. Pharm Res. 2000 Feb;17(2):209-15. doi: 10.1023/a:1007525616017.
|
| 65 |
The influence of oral antidiabetic drugs on cellular drug uptake mediated by hepatic OATP family members. Basic Clin Pharmacol Toxicol. 2013 Apr;112(4):244-50. doi: 10.1111/bcpt.12031. Epub 2012 Dec 6.
|
| 66 |
Functional characterization of SLCO1B1 (OATP-C) variants, SLCO1B1*5, SLCO1B1*15 and SLCO1B1*15+C1007G, by using transient expression systems of HeLa and HEK293 cells. Pharmacogenet Genomics. 2005 Jul;15(7):513-22.
|
| 67 |
Identification of target and pathway of aspirin combined with Lipitor treatment in prostate cancer through integrated bioinformatics analysis. Toxicol Appl Pharmacol. 2022 Oct 1;452:116169. doi: 10.1016/j.taap.2022.116169. Epub 2022 Aug 1.
|
| 68 |
A toxicogenomic approach to drug-induced phospholipidosis: analysis of its induction mechanism and establishment of a novel in vitro screening system. Toxicol Sci. 2005 Feb;83(2):282-92.
|
| 69 |
Transcriptome-based functional classifiers for direct immunotoxicity. Arch Toxicol. 2014 Mar;88(3):673-89.
|
| 70 |
Screening autism-associated environmental factors in differentiating human neural progenitors with fractional factorial design-based transcriptomics. Sci Rep. 2023 Jun 29;13(1):10519. doi: 10.1038/s41598-023-37488-0.
|
| 71 |
Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services.
|
| 72 |
Fluoxetine and reversal of multidrug resistance. Cancer Lett. 2006 Jun 18;237(2):180-7.
|
| 73 |
O-Dealkylation of fluoxetine in relation to CYP2C19 gene dose and involvement of CYP3A4 in human liver microsomes. J Pharmacol Exp Ther. 2002 Jan;300(1):105-11.
|
| 74 |
Clinically significant psychotropic drug-drug interactions in the primary care setting. Curr Psychiatry Rep. 2012 Aug;14(4):376-90.
|
| 75 |
(R)-, (S)-, and racemic fluoxetine N-demethylation by human cytochrome P450 enzymes. Drug Metab Dispos. 2000 Oct;28(10):1187-91.
|
| 76 |
Antidepressant drugs in the elderly--role of the cytochrome P450 2D6. World J Biol Psychiatry. 2003 Apr;4(2):74-80.
|
| 77 |
Binding of CYP2C9 with diverse drugs and its implications for metabolic mechanism. Med Chem. 2009 May;5(3):263-70.
|
| 78 |
Molecular characterization of CYP2B6 substrates. Curr Drug Metab. 2008 Jun;9(5):363-73.
|
| 79 |
Effects of CYP2C19 genotype and CYP2C9 on fluoxetine N-demethylation in human liver microsomes. Acta Pharmacol Sin. 2001 Jan;22(1):85-90.
|
| 80 |
Age-dependent antidepressant pharmacogenomics: polymorphisms of the serotonin transporter and G protein beta3 subunit as predictors of response to fluoxetine and nortriptyline. Int J Neuropsychopharmacol. 2003 Dec;6(4):339-46. doi: 10.1017/S1461145703003663.
|
| 81 |
Mechanism-based inactivation of human cytochrome P4502C8 by drugs in vitro. J Pharmacol Exp Ther. 2004 Dec;311(3):996-1007.
|
| 82 |
Determination of phospholipidosis potential based on gene expression analysis in HepG2 cells. Toxicol Sci. 2007 Mar;96(1):101-14.
|
| 83 |
Inverse agonist and neutral antagonist actions of antidepressants at recombinant and native 5-hydroxytryptamine2C receptors: differential modulatio... Mol Pharmacol. 2008 Mar;73(3):748-57.
|
| 84 |
Effects of selective serotonin reuptake inhibitors on three sex steroids in two versions of the aromatase enzyme inhibition assay and in the H295R cell assay. Toxicol In Vitro. 2015 Oct;29(7):1729-35.
|
| 85 |
Antidepressant drugs activate SREBP and up-regulate cholesterol and fatty acid biosynthesis in human glial cells. Neurosci Lett. 2006 Mar 13;395(3):185-90. doi: 10.1016/j.neulet.2005.10.096. Epub 2005 Dec 1.
|
| 86 |
Fluoxetine inhibits the extracellular signal regulated kinase pathway and suppresses growth of cancer cells. Cancer Biol Ther. 2008 Oct;7(10):1685-93. doi: 10.4161/cbt.7.10.6664. Epub 2008 Oct 22.
|
| 87 |
Serum prolactin levels among outpatients with major depressive disorder during the acute phase of treatment with fluoxetine. J Clin Psychiatry. 2006 Jun;67(6):952-7. doi: 10.4088/jcp.v67n0612.
|
| 88 |
Anti-inflammatory properties of desipramine and fluoxetine. Respir Res. 2007 May 3;8(1):35. doi: 10.1186/1465-9921-8-35.
|
| 89 |
Acid sphingomyelinase-ceramide system mediates effects of antidepressant drugs. Nat Med. 2013 Jul;19(7):934-8. doi: 10.1038/nm.3214. Epub 2013 Jun 16.
|
| 90 |
Systems pharmacological analysis of drugs inducing stevens-johnson syndrome and toxic epidermal necrolysis. Chem Res Toxicol. 2015 May 18;28(5):927-34. doi: 10.1021/tx5005248. Epub 2015 Apr 3.
|
| 91 |
Fluoxetine mediates G0/G1 arrest by inducing functional inhibition of cyclin dependent kinase subunit (CKS)1. Biochem Pharmacol. 2008 May 15;75(10):1924-34. doi: 10.1016/j.bcp.2008.02.013. Epub 2008 Feb 17.
|
| 92 |
A cell-based Rb(+)-flux assay of the Kv1.3 potassium channel. Assay Drug Dev Technol. 2007 Jun;5(3):373-80. doi: 10.1089/adt.2006.004.
|
| 93 |
Downregulation of glucose-6-phosphatase expression contributes to fluoxetine-induced hepatic steatosis. J Appl Toxicol. 2021 Aug;41(8):1232-1240. doi: 10.1002/jat.4109. Epub 2020 Nov 12.
|
| 94 |
Palmitate increases the susceptibility of cells to drug-induced toxicity: an in vitro method to identify drugs with potential contraindications in patients with metabolic disease. Toxicol Sci. 2012 Oct;129(2):346-62. doi: 10.1093/toxsci/kfs208. Epub 2012 Jun 14.
|
| 95 |
Drug-induced long QT syndrome: hERG K+ channel block and disruption of protein trafficking by fluoxetine and norfluoxetine. Br J Pharmacol. 2006 Nov;149(5):481-9. doi: 10.1038/sj.bjp.0706892. Epub 2006 Sep 11.
|
| 96 |
The antidepressants maprotiline and fluoxetine induce Type II autophagic cell death in drug-resistant Burkitt's lymphoma. Int J Cancer. 2011 Apr 1;128(7):1712-23. doi: 10.1002/ijc.25477. Epub 2010 May 25.
|
| 97 |
Association of CYP1A1 and CYP1B1 inhibition in in vitro assays with drug-induced liver injury. J Toxicol Sci. 2021;46(4):167-176. doi: 10.2131/jts.46.167.
|
| 98 |
Establishment of ion channel and ABC transporter assays in 3D-cultured ReNcell VM on a 384-pillar plate for neurotoxicity potential. Toxicol In Vitro. 2022 Aug;82:105375. doi: 10.1016/j.tiv.2022.105375. Epub 2022 May 10.
|
| 99 |
Response to fluoxetine and serotonin 1A receptor (C-1019G) polymorphism in Taiwan Chinese major depressive disorder. Pharmacogenomics J. 2006 Jan-Feb;6(1):27-33. doi: 10.1038/sj.tpj.6500340.
|
| 100 |
Lymphoma cells protected from apoptosis by dysregulated bcl-2 continue to bind annexin V in response to B-cell receptor engagement: a cautionary tale. Leuk Res. 2006 Jan;30(1):77-80. doi: 10.1016/j.leukres.2005.05.018. Epub 2005 Aug 1.
|
| 101 |
Effect of repeated treatment with fluoxetine on tryptophan hydroxylase-2 gene expression in the rat brainstem. Pharmacol Biochem Behav. 2006 Sep;85(1):220-7. doi: 10.1016/j.pbb.2006.08.004. Epub 2006 Sep 18.
|
| 102 |
Association of brain-derived neurotrophic factor genetic Val66Met polymorphism with severity of depression, efficacy of fluoxetine and its side effects in Chinese major depressive patients. Neuropsychobiology. 2010;61(2):71-8. doi: 10.1159/000265132. Epub 2009 Dec 12.
|
|
|
|
|
|
|