| DOT Name |
DOT ID |
UniProt ID |
Mode of Action |
REF |
|
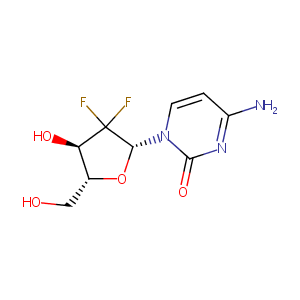
Cytidine deaminase (CDA)
|
OT3HXP6N
|
CDD_HUMAN
|
Affects Response To Substance
|
[15] |
|
ATP-dependent translocase ABCB1 (ABCB1)
|
OTEJROBO
|
MDR1_HUMAN
|
Decreases Expression
|
[25] |
|
Ribonucleoside-diphosphate reductase subunit M2 (RRM2)
|
OTOB6J6R
|
RIR2_HUMAN
|
Affects Response To Substance
|
[16] |
|
Ribonucleoside-diphosphate reductase large subunit (RRM1)
|
OTXGQOR9
|
RIR1_HUMAN
|
Affects Response To Substance
|
[16] |
|
Tumor necrosis factor receptor superfamily member 10B (TNFRSF10B)
|
OTA1CPBV
|
TR10B_HUMAN
|
Increases Expression
|
[26] |
|
Epidermal growth factor receptor (EGFR)
|
OTAPLO1S
|
EGFR_HUMAN
|
Decreases Expression
|
[27] |
|
Apoptosis regulator Bcl-2 (BCL2)
|
OT9DVHC0
|
BCL2_HUMAN
|
Decreases Expression
|
[28] |
|
NF-kappa-B inhibitor epsilon (NFKBIE)
|
OTLAYEL9
|
IKBE_HUMAN
|
Affects Expression
|
[11] |
|
Tumor necrosis factor receptor superfamily member 11B (TNFRSF11B)
|
OTQ4W7MT
|
TR11B_HUMAN
|
Increases Expression
|
[11] |
|
Stearoyl-CoA desaturase (SCD)
|
OTB1073G
|
SCD_HUMAN
|
Increases Expression
|
[11] |
|
1-acyl-sn-glycerol-3-phosphate acyltransferase beta (AGPAT2)
|
OT5I4Y9K
|
PLCB_HUMAN
|
Decreases Expression
|
[11] |
|
Insulin-induced gene 1 protein (INSIG1)
|
OTZF5X1D
|
INSI1_HUMAN
|
Increases Expression
|
[11] |
|
E3 ubiquitin-protein ligase Praja-2 (PJA2)
|
OT45TMC4
|
PJA2_HUMAN
|
Increases Expression
|
[11] |
|
Interleukin-1 receptor-associated kinase-like 2 (IRAK2)
|
OT6Y1QZN
|
IRAK2_HUMAN
|
Increases Expression
|
[11] |
|
Bcl-2-like protein 11 (BCL2L11)
|
OTNQQWFJ
|
B2L11_HUMAN
|
Increases Expression
|
[11] |
|
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3-like (BNIP3L)
|
OTJKOMXE
|
BNI3L_HUMAN
|
Increases Expression
|
[11] |
|
Heterogeneous nuclear ribonucleoprotein Q (SYNCRIP)
|
OTRNDZXB
|
HNRPQ_HUMAN
|
Decreases Expression
|
[11] |
|
Melanoma-associated antigen C1 (MAGEC1)
|
OTHMFHH8
|
MAGC1_HUMAN
|
Increases Expression
|
[11] |
|
Liprin-alpha-4 (PPFIA4)
|
OTWXG0DO
|
LIPA4_HUMAN
|
Increases Expression
|
[11] |
|
Transcription factor Maf (MAF)
|
OT1GR3IZ
|
MAF_HUMAN
|
Increases Expression
|
[11] |
|
Protein phosphatase 1 regulatory subunit 15A (PPP1R15A)
|
OTYG179K
|
PR15A_HUMAN
|
Increases Expression
|
[11] |
|
Krueppel-like factor 8 (KLF8)
|
OTUC5CDB
|
KLF8_HUMAN
|
Increases Expression
|
[11] |
|
Urokinase-type plasminogen activator (PLAU)
|
OTX0QGKK
|
UROK_HUMAN
|
Increases Expression
|
[11] |
|
Fibrinogen gamma chain (FGG)
|
OT5BJSEX
|
FIBG_HUMAN
|
Affects Expression
|
[11] |
|
Interleukin-6 (IL6)
|
OTUOSCCU
|
IL6_HUMAN
|
Increases Expression
|
[11] |
|
Keratin, type II cytoskeletal 8 (KRT8)
|
OTTM4X11
|
K2C8_HUMAN
|
Decreases Expression
|
[11] |
|
Growth-regulated alpha protein (CXCL1)
|
OT3WCTZV
|
GROA_HUMAN
|
Increases Expression
|
[11] |
|
Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1)
|
OTXOQDSG
|
IFIT1_HUMAN
|
Increases Expression
|
[11] |
|
Transcriptional activator Myb (MYB)
|
OTJH64IV
|
MYB_HUMAN
|
Increases Expression
|
[11] |
|
Osteopontin (SPP1)
|
OTJGC23Y
|
OSTP_HUMAN
|
Increases Expression
|
[11] |
|
Endoplasmic reticulum chaperone BiP (HSPA5)
|
OTFUIRAO
|
BIP_HUMAN
|
Increases Expression
|
[11] |
|
Solute carrier family 2, facilitated glucose transporter member 3 (SLC2A3)
|
OT2HZK5M
|
GTR3_HUMAN
|
Increases Expression
|
[11] |
|
Creatine kinase B-type (CKB)
|
OTUCKOTT
|
KCRB_HUMAN
|
Decreases Expression
|
[11] |
|
Cadherin-1 (CDH1)
|
OTFJMXPM
|
CADH1_HUMAN
|
Increases Expression
|
[11] |
|
Homeobox protein Hox-B2 (HOXB2)
|
OTTD6HMV
|
HXB2_HUMAN
|
Increases Expression
|
[11] |
|
Fos-related antigen 2 (FOSL2)
|
OT3MT9HJ
|
FOSL2_HUMAN
|
Decreases Expression
|
[11] |
|
Methylated-DNA--protein-cysteine methyltransferase (MGMT)
|
OT40A9WH
|
MGMT_HUMAN
|
Decreases Expression
|
[11] |
|
CCAAT/enhancer-binding protein beta (CEBPB)
|
OTM9MQIA
|
CEBPB_HUMAN
|
Increases Expression
|
[11] |
|
Early growth response protein 1 (EGR1)
|
OTCP6XGZ
|
EGR1_HUMAN
|
Increases Expression
|
[11] |
|
Cyclic AMP-dependent transcription factor ATF-3 (ATF3)
|
OTC1UOHP
|
ATF3_HUMAN
|
Increases Expression
|
[11] |
|
C-X-C motif chemokine 2 (CXCL2)
|
OTEJCYMY
|
CXCL2_HUMAN
|
Increases Expression
|
[11] |
|
C-X-C motif chemokine 3 (CXCL3)
|
OTSL94KH
|
CXCL3_HUMAN
|
Increases Expression
|
[11] |
|
Endothelin-2 (EDN2)
|
OTQ7RCPI
|
EDN2_HUMAN
|
Decreases Expression
|
[11] |
|
Tumor necrosis factor alpha-induced protein 3 (TNFAIP3)
|
OTVLI4DD
|
TNAP3_HUMAN
|
Increases Expression
|
[11] |
|
Diamine acetyltransferase 1 (SAT1)
|
OT52AU22
|
SAT1_HUMAN
|
Increases Expression
|
[11] |
|
Prostaglandin G/H synthase 1 (PTGS1)
|
OTHCRLEC
|
PGH1_HUMAN
|
Decreases Expression
|
[11] |
|
G1/S-specific cyclin-D1 (CCND1)
|
OT8HPTKJ
|
CCND1_HUMAN
|
Decreases Expression
|
[11] |
|
Tumor necrosis factor receptor superfamily member 5 (CD40)
|
OT5YU2XB
|
TNR5_HUMAN
|
Decreases Expression
|
[11] |
|
Myristoylated alanine-rich C-kinase substrate (MARCKS)
|
OT7N056G
|
MARCS_HUMAN
|
Increases Expression
|
[11] |
|
Protein EVI2B (EVI2B)
|
OT98X1OE
|
EVI2B_HUMAN
|
Decreases Expression
|
[11] |
|
Heat shock 70 kDa protein 1-like (HSPA1L)
|
OTC2V1K6
|
HS71L_HUMAN
|
Decreases Expression
|
[11] |
|
Glutaredoxin-1 (GLRX)
|
OT0QHTAR
|
GLRX1_HUMAN
|
Increases Expression
|
[11] |
|
Cyclin-dependent kinase inhibitor 1 (CDKN1A)
|
OTQWHCZE
|
CDN1A_HUMAN
|
Increases Expression
|
[11] |
|
Nicotinamide N-methyltransferase (NNMT)
|
OTFM38GU
|
NNMT_HUMAN
|
Increases Expression
|
[11] |
|
B-cell lymphoma 6 protein (BCL6)
|
OTQAWWO1
|
BCL6_HUMAN
|
Increases Expression
|
[11] |
|
Regulator of G-protein signaling 2 (RGS2)
|
OT0FSRW7
|
RGS2_HUMAN
|
Increases Expression
|
[11] |
|
Leukemia inhibitory factor receptor (LIFR)
|
OT36W9O5
|
LIFR_HUMAN
|
Increases Expression
|
[11] |
|
Melanoma-associated antigen 10 (MAGEA10)
|
OTI4KPYB
|
MAGAA_HUMAN
|
Decreases Expression
|
[11] |
|
Lysophosphatidic acid receptor 6 (LPAR6)
|
OT4BY3W4
|
LPAR6_HUMAN
|
Decreases Expression
|
[11] |
|
Ras GTPase-activating-like protein IQGAP1 (IQGAP1)
|
OTZRWTGA
|
IQGA1_HUMAN
|
Increases Expression
|
[11] |
|
Retinaldehyde dehydrogenase 3 (ALDH1A3)
|
OT1C9NKQ
|
AL1A3_HUMAN
|
Increases Expression
|
[11] |
|
Max-interacting protein 1 (MXI1)
|
OTUQ9E0D
|
MXI1_HUMAN
|
Increases Expression
|
[11] |
|
ETS translocation variant 1 (ETV1)
|
OT6PMJIK
|
ETV1_HUMAN
|
Increases Expression
|
[11] |
|
Glypican-3 (GPC3)
|
OTWS8WSW
|
GPC3_HUMAN
|
Increases Expression
|
[11] |
|
Transcription initiation factor IIA subunit 1 (GTF2A1)
|
OTWW4PN0
|
TF2AA_HUMAN
|
Affects Expression
|
[11] |
|
Growth arrest-specific protein 1 (GAS1)
|
OTKJXG52
|
GAS1_HUMAN
|
Decreases Expression
|
[11] |
|
Puromycin-sensitive aminopeptidase (NPEPPS)
|
OTMENFXR
|
PSA_HUMAN
|
Decreases Expression
|
[11] |
|
Methionine--tRNA ligase, cytoplasmic (MARS1)
|
OTLJ6MHJ
|
SYMC_HUMAN
|
Increases Expression
|
[11] |
|
Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN (PTEN)
|
OTOWDUNT
|
PTEN_HUMAN
|
Affects Expression
|
[11] |
|
Ras-related protein Rab-8A (RAB8A)
|
OTPB54Y3
|
RAB8A_HUMAN
|
Decreases Expression
|
[11] |
|
Lysozyme C (LYZ)
|
OTD7H0G6
|
LYSC_HUMAN
|
Decreases Expression
|
[11] |
|
Transforming growth factor beta-2 proprotein (TGFB2)
|
OTC0TXEP
|
TGFB2_HUMAN
|
Increases Expression
|
[11] |
|
Large ribosomal subunit protein eL37 (RPL37)
|
OTB6SDZ1
|
RL37_HUMAN
|
Increases Expression
|
[11] |
|
Histone H4 (H4C1)
|
OTB71W46
|
H4_HUMAN
|
Decreases Expression
|
[11] |
|
C-C motif chemokine 20 (CCL20)
|
OTUCJY4N
|
CCL20_HUMAN
|
Increases Expression
|
[11] |
|
ADP-ribosylation factor 1 (ARF1)
|
OT5C4C3L
|
ARF1_HUMAN
|
Decreases Expression
|
[11] |
|
Nuclear factor NF-kappa-B p100 subunit (NFKB2)
|
OTS231V7
|
NFKB2_HUMAN
|
Affects Expression
|
[11] |
|
Methylmalonate-semialdehyde/malonate-semialdehyde dehydrogenase , mitochondrial (ALDH6A1)
|
OT8LCZCT
|
MMSA_HUMAN
|
Increases Expression
|
[11] |
|
Pro-neuregulin-1, membrane-bound isoform (NRG1)
|
OTZO6F1X
|
NRG1_HUMAN
|
Increases Expression
|
[11] |
|
Tyrosine-protein phosphatase non-receptor type 11 (PTPN11)
|
OTFH9M73
|
PTN11_HUMAN
|
Affects Expression
|
[11] |
|
1,25-dihydroxyvitamin D(3) 24-hydroxylase, mitochondrial (CYP24A1)
|
OTG2T749
|
CP24A_HUMAN
|
Decreases Expression
|
[11] |
|
ATP-dependent RNA helicase A (DHX9)
|
OT5AAOQI
|
DHX9_HUMAN
|
Decreases Expression
|
[11] |
|
Nuclear cap-binding protein subunit 1 (NCBP1)
|
OTYBR927
|
NCBP1_HUMAN
|
Increases Expression
|
[11] |
|
TP53-binding protein 1 (TP53BP1)
|
OTL8879S
|
TP53B_HUMAN
|
Increases Expression
|
[11] |
|
Ankyrin-3 (ANK3)
|
OTJ3IRBP
|
ANK3_HUMAN
|
Increases Expression
|
[11] |
|
TNF receptor-associated factor 1 (TRAF1)
|
OTTLM5RU
|
TRAF1_HUMAN
|
Affects Expression
|
[11] |
|
PDZK1-interacting protein 1 (PDZK1IP1)
|
OTWA6M5K
|
PDZ1I_HUMAN
|
Increases Expression
|
[11] |
|
Treacle protein (TCOF1)
|
OT4BOYTM
|
TCOF_HUMAN
|
Decreases Expression
|
[11] |
|
Baculoviral IAP repeat-containing protein 3 (BIRC3)
|
OT3E95KB
|
BIRC3_HUMAN
|
Affects Expression
|
[11] |
|
Baculoviral IAP repeat-containing protein 2 (BIRC2)
|
OTFXFREP
|
BIRC2_HUMAN
|
Increases Expression
|
[11] |
|
Cytoplasmic dynein 1 heavy chain 1 (DYNC1H1)
|
OTD1KRKO
|
DYHC1_HUMAN
|
Decreases Expression
|
[11] |
|
Enhancer of filamentation 1 (NEDD9)
|
OTGCFN4M
|
CASL_HUMAN
|
Increases Expression
|
[11] |
|
Interferon regulatory factor 3 (IRF3)
|
OT81U8ME
|
IRF3_HUMAN
|
Decreases Expression
|
[11] |
|
FXYD domain-containing ion transport regulator 3 (FXYD3)
|
OT9PPRHE
|
FXYD3_HUMAN
|
Increases Expression
|
[11] |
|
Ankyrin repeat domain-containing protein 1 (ANKRD1)
|
OTHJ7JV9
|
ANKR1_HUMAN
|
Increases Expression
|
[11] |
|
Disks large homolog 2 (DLG2)
|
OTQ3BD8U
|
DLG2_HUMAN
|
Increases Expression
|
[11] |
|
Cysteine and glycine-rich protein 2 (CSRP2)
|
OT4SW0HI
|
CSRP2_HUMAN
|
Increases Expression
|
[11] |
|
Cyclin-G2 (CCNG2)
|
OTII38K2
|
CCNG2_HUMAN
|
Increases Expression
|
[11] |
|
Golgi-associated kinase 1B (GASK1B)
|
OT2HPHEI
|
GAK1B_HUMAN
|
Decreases Expression
|
[11] |
|
SUZ domain-containing protein 1 (SZRD1)
|
OTJMEJWT
|
SZRD1_HUMAN
|
Decreases Expression
|
[11] |
|
Leucine-rich repeat-containing protein 28 (LRRC28)
|
OTFTT6VN
|
LRC28_HUMAN
|
Increases Expression
|
[11] |
|
Mitochondrial basic amino acids transporter (SLC25A29)
|
OT09PIFE
|
S2529_HUMAN
|
Increases Expression
|
[11] |
|
Equilibrative nucleobase transporter 1 (SLC43A3)
|
OTR60ZLA
|
S43A3_HUMAN
|
Decreases Expression
|
[11] |
|
Proprotein convertase subtilisin/kexin type 9 (PCSK9)
|
OTI0DU9Y
|
PCSK9_HUMAN
|
Increases Expression
|
[11] |
|
E3 ubiquitin-protein ligase RNF149 (RNF149)
|
OT6SBKQV
|
RN149_HUMAN
|
Increases Expression
|
[11] |
|
F-box only protein 25 (FBXO25)
|
OTDFA7F4
|
FBX25_HUMAN
|
Increases Expression
|
[11] |
|
Solute carrier family 2, facilitated glucose transporter member 14 (SLC2A14)
|
OTBFIOVY
|
GTR14_HUMAN
|
Increases Expression
|
[11] |
|
Membrane progestin receptor beta (PAQR8)
|
OTGZ8W9F
|
PAQR8_HUMAN
|
Increases Expression
|
[11] |
|
Nuclear receptor subfamily 4 group A member 3 (NR4A3)
|
OTPBE9R1
|
NR4A3_HUMAN
|
Increases Expression
|
[11] |
|
La-related protein 4B (LARP4B)
|
OT2HI0QE
|
LAR4B_HUMAN
|
Increases Expression
|
[11] |
|
Collagen triple helix repeat-containing protein 1 (CTHRC1)
|
OTV88X2G
|
CTHR1_HUMAN
|
Increases Expression
|
[11] |
|
BTB/POZ domain-containing protein KCTD12 (KCTD12)
|
OTPYKT4Z
|
KCD12_HUMAN
|
Decreases Expression
|
[11] |
|
Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 (MAGI1)
|
OTMV4ASV
|
MAGI1_HUMAN
|
Increases Expression
|
[11] |
|
Sialidase-1 (NEU1)
|
OTH9BY8Y
|
NEUR1_HUMAN
|
Increases Expression
|
[11] |
|
Galactosylceramide sulfotransferase (GAL3ST1)
|
OTSFFZRD
|
G3ST1_HUMAN
|
Decreases Expression
|
[11] |
|
Junctional adhesion molecule C (JAM3)
|
OTX0F9QL
|
JAM3_HUMAN
|
Decreases Expression
|
[11] |
|
SLIT and NTRK-like protein 2 (SLITRK2)
|
OTPVI22J
|
SLIK2_HUMAN
|
Increases Expression
|
[11] |
|
Anaphase-promoting complex subunit 1 (ANAPC1)
|
OT2YY35L
|
APC1_HUMAN
|
Increases Expression
|
[11] |
|
Anthrax toxin receptor 1 (ANTXR1)
|
OT5W1GPC
|
ANTR1_HUMAN
|
Affects Expression
|
[11] |
|
Ras-related GTP-binding protein D (RRAGD)
|
OTXLVWAH
|
RRAGD_HUMAN
|
Increases Expression
|
[11] |
|
DNA damage-inducible transcript 4 protein (DDIT4)
|
OTHY8SY4
|
DDIT4_HUMAN
|
Affects Expression
|
[11] |
|
Intersectin-2 (ITSN2)
|
OT8S0OO8
|
ITSN2_HUMAN
|
Increases Expression
|
[11] |
|
Probable ribosome biogenesis protein RLP24 (RSL24D1)
|
OT6KI56F
|
RLP24_HUMAN
|
Increases Expression
|
[11] |
|
Transcription elongation factor A protein-like 9 (TCEAL9)
|
OT4UHDB5
|
TCAL9_HUMAN
|
Increases Expression
|
[11] |
|
Histone deacetylase 9 (HDAC9)
|
OTO8O0LF
|
HDAC9_HUMAN
|
Increases Expression
|
[11] |
|
Phosphoserine aminotransferase (PSAT1)
|
OTVV1YV9
|
SERC_HUMAN
|
Increases Expression
|
[11] |
|
Chloride intracellular channel protein 4 (CLIC4)
|
OT6KTPKD
|
CLIC4_HUMAN
|
Increases Expression
|
[11] |
|
Alpha-2-macroglobulin (A2M)
|
OTFTX90K
|
A2MG_HUMAN
|
Increases Expression
|
[12] |
|
Pro-epidermal growth factor (EGF)
|
OTANRJ0L
|
EGF_HUMAN
|
Decreases Expression
|
[12] |
|
Insulin (INS)
|
OTZ85PDU
|
INS_HUMAN
|
Increases Expression
|
[12] |
|
Lymphotoxin-alpha (LTA)
|
OTUC3S76
|
TNFB_HUMAN
|
Increases Expression
|
[12] |
|
Tumor necrosis factor (TNF)
|
OT4IE164
|
TNFA_HUMAN
|
Increases Expression
|
[12] |
|
Interferon gamma (IFNG)
|
OTXG9JM7
|
IFNG_HUMAN
|
Decreases Expression
|
[12] |
|
Interleukin-1 alpha (IL1A)
|
OTPSGILV
|
IL1A_HUMAN
|
Decreases Expression
|
[12] |
|
Interleukin-1 beta (IL1B)
|
OT0DWXXB
|
IL1B_HUMAN
|
Decreases Expression
|
[12] |
|
Serum amyloid P-component (APCS)
|
OT76JZ8Z
|
SAMP_HUMAN
|
Decreases Expression
|
[12] |
|
Beta-2-glycoprotein 1 (APOH)
|
OTFMUX6O
|
APOH_HUMAN
|
Increases Expression
|
[12] |
|
Alpha-fetoprotein (AFP)
|
OT9GG3ZI
|
FETA_HUMAN
|
Decreases Expression
|
[12] |
|
Granulocyte-macrophage colony-stimulating factor (CSF2)
|
OT1M7D28
|
CSF2_HUMAN
|
Decreases Expression
|
[12] |
|
Interleukin-4 (IL4)
|
OTOXBWAU
|
IL4_HUMAN
|
Decreases Expression
|
[12] |
|
Myeloperoxidase (MPO)
|
OTOOXLIN
|
PERM_HUMAN
|
Increases Expression
|
[12] |
|
Endothelin-1 (EDN1)
|
OTZCACEG
|
EDN1_HUMAN
|
Decreases Expression
|
[12] |
|
Intercellular adhesion molecule 1 (ICAM1)
|
OTTOIX77
|
ICAM1_HUMAN
|
Decreases Expression
|
[12] |
|
Stromelysin-1 (MMP3)
|
OTGBI74Z
|
MMP3_HUMAN
|
Decreases Expression
|
[12] |
|
Coagulation factor VII (F7)
|
OTGNJ97M
|
FA7_HUMAN
|
Decreases Expression
|
[12] |
|
Fibroblast growth factor 2 (FGF2)
|
OT7YUJ9F
|
FGF2_HUMAN
|
Increases Expression
|
[12] |
|
Granulocyte colony-stimulating factor (CSF3)
|
OT9GC6TP
|
CSF3_HUMAN
|
Decreases Expression
|
[12] |
|
Interleukin-8 (CXCL8)
|
OTS7T5VH
|
IL8_HUMAN
|
Decreases Expression
|
[12] |
|
C-C motif chemokine 3 (CCL3)
|
OTW2H3ND
|
CCL3_HUMAN
|
Increases Expression
|
[12] |
|
C-C motif chemokine 2 (CCL2)
|
OTAD2HEL
|
CCL2_HUMAN
|
Decreases Expression
|
[12] |
|
Prostatic acid phosphatase (ACP3)
|
OTSIM0YG
|
PPAP_HUMAN
|
Decreases Expression
|
[12] |
|
Kit ligand (KITLG)
|
OTB9AVQ4
|
SCF_HUMAN
|
Decreases Expression
|
[12] |
|
Interleukin-10 (IL10)
|
OTIRFRXC
|
IL10_HUMAN
|
Decreases Expression
|
[12] |
|
Brain-derived neurotrophic factor (BDNF)
|
OTLGH7EW
|
BDNF_HUMAN
|
Decreases Expression
|
[12] |
|
CD40 ligand (CD40LG)
|
OT75Z6A6
|
CD40L_HUMAN
|
Decreases Expression
|
[12] |
|
Interleukin-13 (IL13)
|
OTI4YS3Y
|
IL13_HUMAN
|
Increases Expression
|
[12] |
|
Thrombopoietin (THPO)
|
OTO73DZ2
|
TPO_HUMAN
|
Increases Expression
|
[12] |
|
Interleukin-15 (IL15)
|
OTQSYWQS
|
IL15_HUMAN
|
Decreases Expression
|
[12] |
|
Leptin (LEP)
|
OT5Q7ODW
|
LEP_HUMAN
|
Decreases Expression
|
[12] |
|
Pappalysin-1 (PAPPA)
|
OTTTG9PG
|
PAPP1_HUMAN
|
Decreases Expression
|
[12] |
|
Pro-interleukin-16 (IL16)
|
OTZ2MO6B
|
IL16_HUMAN
|
Increases Expression
|
[12] |
|
Adiponectin (ADIPOQ)
|
OTNX23LE
|
ADIPO_HUMAN
|
Increases Expression
|
[12] |
|
Aldehyde dehydrogenase 1A1 (ALDH1A1)
|
OTCUWZKB
|
AL1A1_HUMAN
|
Decreases Expression
|
[29] |
|
Neurogenic locus notch homolog protein 1 (NOTCH1)
|
OTI1WADQ
|
NOTC1_HUMAN
|
Increases Expression
|
[29] |
|
Proto-oncogene c-Rel (REL)
|
OTTCKMAC
|
REL_HUMAN
|
Decreases Expression
|
[29] |
|
Telomerase reverse transcriptase (TERT)
|
OT085VVA
|
TERT_HUMAN
|
Decreases Activity
|
[30] |
|
Baculoviral IAP repeat-containing protein 5 (BIRC5)
|
OTILXZYL
|
BIRC5_HUMAN
|
Decreases Expression
|
[31] |
|
Nuclear protein 1 (NUPR1)
|
OT4FU8C0
|
NUPR1_HUMAN
|
Decreases Expression
|
[32] |
|
Collagen alpha-1(I) chain (COL1A1)
|
OTI31178
|
CO1A1_HUMAN
|
Decreases Expression
|
[33] |
|
Cellular tumor antigen p53 (TP53)
|
OTIE1VH3
|
P53_HUMAN
|
Increases Expression
|
[34] |
|
Insulin-like growth factor 1 receptor (IGF1R)
|
OTXJIF13
|
IGF1R_HUMAN
|
Decreases Expression
|
[27] |
|
Heme oxygenase 1 (HMOX1)
|
OTC1W6UX
|
HMOX1_HUMAN
|
Increases Expression
|
[35] |
|
Poly polymerase 1 (PARP1)
|
OT310QSG
|
PARP1_HUMAN
|
Increases Cleavage
|
[36] |
|
Zinc finger protein GLI2 (GLI2)
|
OTIRV97L
|
GLI2_HUMAN
|
Affects Expression
|
[27] |
|
Clusterin (CLU)
|
OTQGG0JM
|
CLUS_HUMAN
|
Increases Expression
|
[37] |
|
G2/mitotic-specific cyclin-B1 (CCNB1)
|
OT19S7E5
|
CCNB1_HUMAN
|
Decreases Expression
|
[27] |
|
Serine/threonine-protein kinase B-raf (BRAF)
|
OT7S81XQ
|
BRAF_HUMAN
|
Decreases Expression
|
[27] |
|
NAD(P)H dehydrogenase 1 (NQO1)
|
OTZGGIVK
|
NQO1_HUMAN
|
Increases Expression
|
[35] |
|
CD44 antigen (CD44)
|
OT9TTJ41
|
CD44_HUMAN
|
Affects Expression
|
[25] |
|
Histone H2AX (H2AX)
|
OT18UX57
|
H2AX_HUMAN
|
Increases Expression
|
[38] |
|
Insulin-like growth factor-binding protein 3 (IGFBP3)
|
OTIX63TX
|
IBP3_HUMAN
|
Affects Expression
|
[27] |
|
Cannabinoid receptor 1 (CNR1)
|
OTEALO6G
|
CNR1_HUMAN
|
Increases Expression
|
[39] |
|
Tumor necrosis factor receptor superfamily member 6 (FAS)
|
OTP9XG86
|
TNR6_HUMAN
|
Increases Expression
|
[40] |
|
Mitogen-activated protein kinase 3 (MAPK3)
|
OTCYKGKO
|
MK03_HUMAN
|
Increases Phosphorylation
|
[28] |
|
Aryl hydrocarbon receptor nuclear translocator (ARNT)
|
OTMSIEZY
|
ARNT_HUMAN
|
Increases Expression
|
[27] |
|
Mitogen-activated protein kinase 1 (MAPK1)
|
OTH85PI5
|
MK01_HUMAN
|
Increases Phosphorylation
|
[28] |
|
14-3-3 protein sigma (SFN)
|
OTLJCZ1U
|
1433S_HUMAN
|
Increases Methylation
|
[41] |
|
Cannabinoid receptor 2 (CNR2)
|
OTYP9P43
|
CNR2_HUMAN
|
Increases Expression
|
[39] |
|
Catenin beta-1 (CTNNB1)
|
OTZ932A3
|
CTNB1_HUMAN
|
Increases Expression
|
[42] |
|
Breast cancer type 1 susceptibility protein (BRCA1)
|
OT5BN6VH
|
BRCA1_HUMAN
|
Increases Expression
|
[27] |
|
Signal transducer and activator of transcription 3 (STAT3)
|
OTAAGKYZ
|
STAT3_HUMAN
|
Decreases Activity
|
[43] |
|
Caspase-3 (CASP3)
|
OTIJRBE7
|
CASP3_HUMAN
|
Increases Activity
|
[44] |
|
Proliferation marker protein Ki-67 (MKI67)
|
OTA8N1QI
|
KI67_HUMAN
|
Decreases Expression
|
[31] |
|
Dual specificity mitogen-activated protein kinase kinase 3 (MAP2K3)
|
OTI2OREX
|
MP2K3_HUMAN
|
Increases Phosphorylation
|
[45] |
|
Transcription factor SOX-2 (SOX2)
|
OTFAWCAU
|
SOX2_HUMAN
|
Increases Expression
|
[42] |
|
Glycogen synthase kinase-3 beta (GSK3B)
|
OTL3L14B
|
GSK3B_HUMAN
|
Increases Phosphorylation
|
[46] |
|
Dual specificity mitogen-activated protein kinase kinase 6 (MAP2K6)
|
OTK13JKC
|
MP2K6_HUMAN
|
Increases Phosphorylation
|
[45] |
|
Serine/threonine-protein kinase PLK1 (PLK1)
|
OTRZX45T
|
PLK1_HUMAN
|
Decreases Expression
|
[27] |
|
Caspase-9 (CASP9)
|
OTD4RFFG
|
CASP9_HUMAN
|
Increases Activity
|
[44] |
|
BH3-interacting domain death agonist (BID)
|
OTOSHSHU
|
BID_HUMAN
|
Increases Cleavage
|
[47] |
|
C-X-C chemokine receptor type 4 (CXCR4)
|
OTUFSBX2
|
CXCR4_HUMAN
|
Affects Expression
|
[27] |
|
E3 ubiquitin-protein ligase Mdm2 (MDM2)
|
OTOVXARF
|
MDM2_HUMAN
|
Decreases Expression
|
[27] |
|
POU domain, class 5, transcription factor 1 (POU5F1)
|
OTDHHN7O
|
PO5F1_HUMAN
|
Decreases Expression
|
[25] |
|
Transcription factor p65 (RELA)
|
OTUJP9CN
|
TF65_HUMAN
|
Increases Expression
|
[48] |
|
Regenerating islet-derived protein 3-alpha (REG3A)
|
OT0BVQRB
|
REG3A_HUMAN
|
Decreases Expression
|
[46] |
|
Bcl-2-like protein 1 (BCL2L1)
|
OTRC5K9O
|
B2CL1_HUMAN
|
Decreases Expression
|
[28] |
|
Protein patched homolog 1 (PTCH1)
|
OTMG07H5
|
PTC1_HUMAN
|
Affects Expression
|
[27] |
|
Caspase-8 (CASP8)
|
OTA8TVI8
|
CASP8_HUMAN
|
Increases Activity
|
[44] |
|
Sonic hedgehog protein (SHH)
|
OTOG2BXF
|
SHH_HUMAN
|
Decreases Expression
|
[27] |
|
Nuclear factor erythroid 2-related factor 2 (NFE2L2)
|
OT0HENJ5
|
NF2L2_HUMAN
|
Increases Expression
|
[35] |
|
Tumor protein p53-inducible nuclear protein 1 (TP53INP1)
|
OT2363Z9
|
T53I1_HUMAN
|
Increases Expression
|
[46] |
|
Protein smoothened (SMO)
|
OTXXE208
|
SMO_HUMAN
|
Increases Expression
|
[42] |
|
Homeobox protein NANOG (NANOG)
|
OTUEY1FM
|
NANOG_HUMAN
|
Decreases Expression
|
[42] |
|
Axin-2 (AXIN2)
|
OTRMGQNU
|
AXIN2_HUMAN
|
Affects Expression
|
[27] |
|
3',5'-cyclic-AMP phosphodiesterase 4D (PDE4D)
|
OT1RWFV0
|
PDE4D_HUMAN
|
Increases ADR
|
[49] |
|
Focal adhesion kinase 1 (PTK2)
|
OT3Q1JDY
|
FAK1_HUMAN
|
Decreases Response To Substance
|
[50] |
|
RAC-alpha serine/threonine-protein kinase (AKT1)
|
OT8H2YY7
|
AKT1_HUMAN
|
Affects Response To Substance
|
[51] |
|
Splicing factor 45 (RBM17)
|
OT9ROJCL
|
SPF45_HUMAN
|
Decreases Response To Substance
|
[52] |
|
Hypoxia-inducible factor 1-alpha (HIF1A)
|
OTADSC03
|
HIF1A_HUMAN
|
Affects Response To Substance
|
[53] |
|
Neuropilin-1 (NRP1)
|
OTCGULYV
|
NRP1_HUMAN
|
Decreases Response To Substance
|
[54] |
|
Oxysterol-binding protein 2 (OSBP2)
|
OTDGLB4J
|
OSBP2_HUMAN
|
Increases ADR
|
[49] |
|
Proto-oncogene tyrosine-protein kinase Src (SRC)
|
OTETYX40
|
SRC_HUMAN
|
Decreases Response To Substance
|
[55] |
|
Equilibrative nucleoside transporter 3 (SLC29A3)
|
OTGLX8XU
|
S29A3_HUMAN
|
Affects Transport
|
[56] |
|
Probable ATP-dependent RNA helicase DDX53 (DDX53)
|
OTHK3EGZ
|
DDX53_HUMAN
|
Affects Response To Substance
|
[57] |
|
Amphiregulin (AREG)
|
OTJFOR67
|
AREG_HUMAN
|
Decreases Response To Substance
|
[58] |
|
Pyruvate kinase PKM (PKM)
|
OTLHHMC2
|
KPYM_HUMAN
|
Affects Response To Substance
|
[31] |
|
Equilibrative nucleoside transporter 1 (SLC29A1)
|
OTLOOZZS
|
S29A1_HUMAN
|
Increases Response To Substance
|
[59] |
|
Alpha-protein kinase 3 (ALPK3)
|
OTLUYSMO
|
ALPK3_HUMAN
|
Increases ADR
|
[49] |
|
Transcription factor AP-2-alpha (TFAP2A)
|
OTMYT3NK
|
AP2A_HUMAN
|
Increases Response To Substance
|
[60] |
|
Death-associated protein kinase 1 (DAPK1)
|
OTNCNUCO
|
DAPK1_HUMAN
|
Increases ADR
|
[61] |
|
Cytochrome c oxidase subunit 3 (COX3)
|
OTNNGBYJ
|
COX3_HUMAN
|
Decreases Response To Substance
|
[62] |
|
Cyclin-dependent kinase inhibitor 1B (CDKN1B)
|
OTNY5LLZ
|
CDN1B_HUMAN
|
Decreases Response To Substance
|
[63] |
|
Receptor tyrosine-protein kinase erbB-2 (ERBB2)
|
OTOAUNCK
|
ERBB2_HUMAN
|
Increases ADR
|
[64] |
|
Myc proto-oncogene protein (MYC)
|
OTPV5LUK
|
MYC_HUMAN
|
Affects Response To Substance
|
[65] |
|
3-phosphoinositide-dependent protein kinase 1 (PDPK1)
|
OTT09ZVP
|
PDPK1_HUMAN
|
Decreases Response To Substance
|
[66] |
|
Nucleophosmin (NPM1)
|
OTTBYYT0
|
NPM_HUMAN
|
Decreases Response To Substance
|
[67] |
|
Serine/threonine-protein kinase SMG1 (SMG1)
|
OTTS3SXE
|
SMG1_HUMAN
|
Decreases Response To Substance
|
[68] |
|
ATP-dependent DNA helicase PIF1 (PIF1)
|
OTUHKKVP
|
PIF1_HUMAN
|
Decreases Response To Substance
|
[38] |
|
Transforming growth factor beta-1 proprotein (TGFB1)
|
OTV5XHVH
|
TGFB1_HUMAN
|
Decreases Response To Substance
|
[69] |
|
Ribonucleoside-diphosphate reductase large subunit (RRM1)
|
OTXGQOR9
|
RIR1_HUMAN
|
Increases ADR
|
[70] |
|
Olfactory receptor 4D6 (OR4D6)
|
OTXJKUT1
|
OR4D6_HUMAN
|
Affects Response To Substance
|
[57] |
|
PDZ domain-containing protein GIPC1 (GIPC1)
|
OTXLVCPJ
|
GIPC1_HUMAN
|
Affects Response To Substance
|
[71] |
|
Integrin-linked protein kinase (ILK)
|
OTYG2FD1
|
ILK_HUMAN
|
Affects Response To Substance
|
[51] |
|
Poly polymerase 2 (PARP2)
|
OTYL81ZI
|
PARP2_HUMAN
|
Decreases Response To Substance
|
[72] |
| ------------------------------------------------------------------------------------ |
|
|
|
|